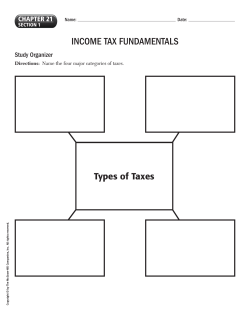
Genomics
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chapter 16 Microbial Genomics 1 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. March 24, 2015 微免科 陳怡原 [email protected] Ext. 3352 2 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Today’s lecture will discuss: 1. 2. 3. 4. 5. What is bacterial genome? How to determine and analyze the genome? What is proteome? How to determine the proteome? What can we learn from the organization and function of a bacterial genome? 3 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Genomics • study of molecular organization of genomes, their information content, and gene products they encode – network of interconnected circuits – window into entire microbial communities – evolutionary insights 4 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Determining DNA Sequences • Sanger DNA sequencing – most commonly used method – referred to as the chain-termination DNA sequencing method – uses dideoxynucleoside triphosphates (ddNTP) 5 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Sanger Method Figure 16.2 6 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Sanger Method • mix single strands of DNA with primer, DNA polymerase I, 4 deoxynucleotides (one of which is radiolabeled), small amount of one ddNTP • DNA synthesis occurs; random insertion of ddNTP generates DNA fragments of different lengths • four reactions carried out; each with different ddNTP • fragments in each reaction mixture separated electrophoretically • gel autoradiographed and sequence read 7 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. More Sanger Sequencing… • automated sequencing – uses four different fluorescent color dyes instead of radiolabeled ddNTP – electrophoresis and laser beam determines order 8 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. The Automated Sanger DNA Sequencing Figure 16.3 9 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Post-Sanger DNA Sequencing • search for more rapid, less cumbersome, and less expensive methods • methods – pyrosequencing – SOLEXA sequencing – SOLiD technology 10 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Genome Sequencing • Whole-genome shotgun sequencing • Single cell genome sequencing 11 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Whole-Genome Shotgun Sequencing • developed in 1995 by J. Craig Venter and Hamilton Smith • four stage process – library construction • generates clones of portions of genome – random sequencing • determines sequences of clones – fragment alignment and gap closure – editing 12 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Whole-Genome Shotgun Sequencing Figure 16.6 13 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Single-Cell Genomic Sequencing • femtograms of DNA from a single cell are amplified to micrograms of DNA needed for sequencing • multiple strand displacement (MSD) – amplification of 70–75% of genome amplified – combining cells reduces mistakes 14 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Single-Cell Genomic Sequencing Figure 16.7 15 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Bioinformatics • analysis of genome data using computers • generates data on genome content, structure, and arrangement • also provides data on protein structure and function • uses annotation to determine location of genes on newly sequenced genome • further examination carried out using in silico analysis 16 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Genome Annotation - 1 • process that locates genes in the genome map • identifies each open reading frame in genome – a reading frame >100 codons that is not interrupted by a stop codon – there is an apparent ribosomal binding site at the 5’ end and terminator sequences at the 3’ end 17 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Finding Potential Protein Coding Genes Figure 16.8 18 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Bioinformatics - 2 • ORFs are presumed to encode protein (coding sequences, CDS) • BLAST (basic local alignment search tool) computer program – base by base comparison of two or more gene sequences – assign tentative function of gene or protein structure 19 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Analysis of Conserved Regions Figure 16.9 20 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Functional Genomics • determination of how genome works • uses physical maps of location of genes • provides information on – metabolic pathways – transport mechanisms – regulatory and signal transduction mechanisms 21 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Physical Map of the Haemophilus influenza Genome Figure 16.10 22 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Metabolic Pathways and Transport systems of Treponema pallidum Figure 16.11 23 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. DNA Microarray Analysis • can determine which genes are expressed at a specific time • arrays are solid supports to which DNA is attached • each DNA spot (called a probe), represents a single gene or ORF • commercially prepared microarrays can be purchased 24 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Gene Microarrays • spotted arrays – prepared by robotic application of DNA probe • examples of probes = PCR product, cDNA or oligonucleotide) • oligonucleotide probes from eukaryotes are called expressed sequence tags (EST) 25 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Analysis of Gene Expression Using Microarrays • based on hybridization between the probe DNA and the targets (the nucleic acids to be analyzed) – targets are labeled with fluorescent dyes and then incubated with the gene chip – unbound target is washed off – the chip is scanned with laser beams to detect fluorescence which indicates that hybridization has occurred 26 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. A Microarray System Figure 16.12 27 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Applications of Microarray Analysis • which genes have changed expression in response to environmental changes – hierarchical cluster analysis • groups genes with similar function or patterns of regulation – analysis of transcriptome - all the mRNA present after environmental challenge 28 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Hierarchical Cluster Analysis of Gene Expression Figure 16.13 29 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Proteomics • the study of the proteome – the entire collection of proteins that an organism produces • provides information about genome function not available from mRNA studies • information determines what is actually happening in cells is referred to as functional proteomics 30 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Analysis of Proteome • proteome often analyzed by two-dimensional gel electrophoresis – first dimension • isoelectric focusing – pH gradient determines isoelectric point – second dimension • electrophoresis (SDSPAGE) and separation by molecular weight Figure 16.14 31 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Further Proteome Analysis • Tandem Mass Spectrometry – unknown spot from 2-D gel is cut and cleaved – fragments are analyzed by mass spectrometer – mass of fragments is plotted – protein tentatively identified from probable amino acid composition 32 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Mass Spectrometry Figure 16.15 33 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Structural Proteomics • protein modeling – determination of 3-D structure of proteins – predicting structure of other proteins and proteins complexes • lipidomics, glycomics – cell’s lipid and carbohydrate profile • metabolomics – small molecule metabolites 34 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Probing DNA-Protein Interactions • chromatin immunoprecipitation (ChIP) – allows for study of interactions in absence of purified protein – proteins are stably attached to DNA – protein specific antibodies are cross-linked – fluorescently labeled DNA is hybridized to microarray for identification 35 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. ChIP-Chip Analysis Figure 16.16 36 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Systems Biology • integration of “parts lists” of cells with molecular interactions that become pathways for catabolism, anabolism, regulation, behavior, environmental responses, etc. – holistic study of cells – may be important in studying host microbe interactions 37 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Comparative Genomics - 1 • set of analyses by which gene function and evolution can be inferred by studying similar nucleotide and amino acid sequences found among organisms • comparisons within domains 38 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Microbial Genome Sizes Figure 16.17 39 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Comparative Genomics - 2 • comparisons of genomes of strains within species and among species – core genome • set of genes found in all members of a monophyletic group • represent minimal number of genes needed – pan-genome • combination of all genes of all strains 40 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Core and Pan-Genomes Figure 16.18 41 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Comparative Genomics - 3 • horizontal gene transfer (HGT) • genomic islands (permanently integrated mobile genetic elements) • pathogenicity islands (virulence proteins) • phylogenic relationships between microbes can be studied by synteny – order of genes on genome 42 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Synteny Figure 16.19 43 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Comparative Genomics - 4 • pathogen virulence genes – pseudogenes – non-functional genes in non-pathogen relatives • reverse vaccinology – development of new vaccines using only specific proteins of pathogens 44 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Comparative Genomic Analysis between two Mycobacterium species Figure 16.20 45 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Metagenomics • environmental genomics – cultivation-independent technique – used to learn more about the diversity and metabolic potential of microbial communities – takes a census of microbial populations and can determine the presence and level of classes of genes 46 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. From the environment…… Figure 16.21 47 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Phylotypes • genes that indicate taxonomy may aid in metabolic activities of microbial communities • examples – finding rhodopsin-like genes in marine microbes is an exciting discovery that requires a reassessment of oceanic carbon cycles – prokaryotic biodiversity of Segrasso sea – microbiome in humans and normal microbiota 48 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Phylogenetic Diversity of Sargasso Sea Microbes Figure 16.22 49 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Summary • • • • • • • Genome sequencing Bioinformatics Functional genomics Proteomics System biology Comparative genomics Metagenomics 50
© Copyright 2026