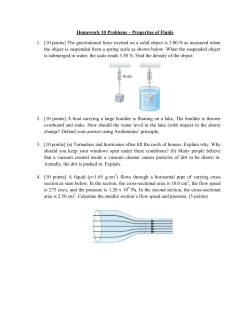
The Protein Interaction Prediction Engine (PIPE)
The Protein Interaction Prediction Engine (PIPE) Project Frank Dehne School of Computer Science Carleton University Ashkan Golshani Institute for Biochemistry Carleton University PIPE Project Project started in 2003 Multi-disciplinary team: Computer Science – F.Dehne Biochemistry – A.Golshani – M.Dumontier – J.Greenblatt Biomed. Eng. – J. Green Graduate Students: S.Pitre, C.North, A.Amos-Binks, A. Schoenrock, M.Alamgir, Bahram Samanfar, Mohsen Hooshyar, ... Equipment: 256 core PC Cluster 1168 core Sun T2 “Victoria Falls” Cluster 32,000 core Blue Gene/Q Basics of Proteins Why do we study protein-protein interactions (PPIs)? 1.Proteins often perform their function(s) by interacting with other proteins For example: RNA pol II 1. PPIs form a major group of interaction networks within a cell i.e. help define the biology of a cell as a system. 3rd yr Biology undergrads Biology Admin staff Biology Profs Biology TA Conclusion: He is most likely a biology student. Nuclear Export Variant Histone Ubuiquination TREX CPF Mediator COMPASS ER to Golgi Transport Elongator RNAP II Biochemical Methods Tandem affinity purification (TAP) Yeast two-hybrid screen Co-immunoprecipitation Bimolecular fluorescence complementation Affinity electrophoresis Pull-down assays Label transfer Phage display In-vivo crosslinking Chemical cross-linking Strepprotein interaction experiment Frank Dehne ● www.dehne.net Quantitative immunoprecipitation combined with Tandem Affinity Purification (TAP) Do YGL227W and YMR135C interact? Frank Dehne ● www.dehne.net TAP tag Y2H Computation-based methods may offer improved alternatives. Can we detect PPI based on primary sequence only? - No pre-defined information - Can be applied to all proteins - Can be applied to all genomes, even newly sequenced ones Working Hypothesis Certain regions of interactions can be very small (20 - 40 amino acids) Interaction “Codes” Known Protein Interactions species # proteins # protein pairs # known interactions # unknown interactions S. cerevisiae 6,300 19,867,056 15,151 ??? C. elegans 23,684 280,454,086 6,607 ??? H. sapiens 22,513 253,406,328 41,678 ??? S. cerevisiae C. elegans H. sapiens Basic PIPE Algorithm String comparison Match = (Sum of pairwise PAM values > Threshold) PIPE Output PIPE Output PIPE: Detecting Novel Protein-Protein Interactions S. cerevisiae Banting and Best Institute of Medical Research, Toronto Protein complex: YGL227W, YMR135C, YIL017C, YDL176W, YIL097W, YDR255C, YBR105C PIPE: Elucidating the Architecture of Protein Complexes S. cerevisiae PIPE: Reducing False Positives Eliminate “popular motifs” via median filter PIPE's Prediction Accuracy Many Other Methods... Types of Protein Interaction Prediction Methods: Phylogenetic profiling Identification of homologous interacting pairs Identification of structural patterns (Van der Waals) Bayesian network modelling 3D template-based protein complex modelling Supervised learning (SVM) Park's Comparison Experiment Park (BMC Bioinformatics, 2009, 10:419) compared the four best methods [M1] Martin et.al. (Bioinformatics 2005,21(2):218-226): protein pair is encoded by a product of signatures which is then classified by a support vector classifier [M2] PIPE [M3] Shen et.al. (Proc Natl Acad Sci USA 2007, 104(11):4337-4341): each protein sequence is encoded by a feature vector that represents the frequencies of 3 amino acid-long subsequences, and feature vectors are concatenated for a pair of proteins and classified by SVM. [M4] Guo et.al. (Nucl Acids Res 2008,36(9):3025-3030): each protein sequence is encoded by a feature vector that represents auto-correlation values of 7 different physicochemical scales, and feature vectors are concatenated for a pair of proteins and classified by SVM. Consensus Method: “Vote” among M1-M4. Park's Comparison Experiment From: Park, BMC Bioinformatics, 2009, 10:419 Global Scan of Entire Protein Interaction Network species # proteins # protein pairs # known interactions S. cerevisiae 6,300 19,867,056 15,151 C. elegans 23,684 280,454,08 6 6,607 H. sapiens 22,513 253,406,32 8 41,678 Open Problem... Challenges: Large number of protein pairs (requires high speed, SVM not possible) Small number of true positives (very sparse, ~ 0.1 % density) Requires very high specificity ~99.95 % (i.e. less than 0.05% false positive rate) – Otherwise: #false positives > #true positives PIPE's Prediction Accuracy PIPE Sequential Performance Improvements Character based amino acid representation was converted into binary encodings. Removed need for character-to-index lookup in PAM120. “Sliding window” process was improved to use incremental updates. Pre-computed all possible protein fragment comparisons and stored all matches of similar fragments in a hash table. Large Scale Parallelization: MP-PIPE Architecture: Cluster of multi-core processors One MP-PIPE worker per proc. Each worker with multiple threads H.sapiens protein pairs Summary of PIPE Results PIPE's superior performance and prediction accuracy enabled the first ever complete scan of entire protein interaction networks species # proteins # protein pairs # known interactions # novel PIPE pred. * S. cerevisiae 6,300 19,867,056 15,151 14,438 1 hour C. elegans 23,684 280,454,08 6 6,607 32,548 1 week H.sapiens 22,513 253,406,32 8 41,678 130,470 3 months * False positive rate: 0.0001 256 core PC Cluster 1168 core Sun T2 “Victoria Falls” Cluster Running time Listens from overall PPI profile Previously reported PPIs Novel PPIs (current study) Profile of PPI pairs on the basis of cellular process Babu et al Nature 489: 585-589 H.sapiens dsDNA break repair l Blue: Proteins known to be involved in dsDNA break repair Green: Known interaction Red: Novel interactions discovered by PIPE Yellow: Novel proteins likely involved in dsDNA break repair Frank Dehne ● www.dehne.net Online Resources InSiPS: In-Silico Protein Synthesizer Participants: l l Frank Dehne, Computer Science (PI) Andrew Schoenrock, Computer Science (grad.stud.) Sylavin Pitre, Computer Science (postdoc) Ashkan Golshani, Biochemistry (Co-PI) Dan Burnside, Biochemistry (grad.stud.) Houman Moteshareie, Biochemistry (grad.stud.) Frank Dehne ● www.dehne.net In Silico Protein Synthesizer (InSiPS) The In Silico Protein Synthesizer (InSiPS) is a computational tool that can synthesize proteins with specific protein-protein interaction prediction profiles. Given a set of target proteins and a set of non-target proteins, InSiPS can generate a protein sequence that is predicted to interact with the target proteins and predicted not to interact with the non-targets. Frank Dehne ● www.dehne.net Frank Dehne ● www.dehne.net Fitness Function Frank Dehne ● www.dehne.net Performance on BGQ Frank Dehne ● www.dehne.net Performance on BGQ Frank Dehne ● www.dehne.net Target Proteins (Yeast) Frank Dehne ● www.dehne.net Parameter Settings Frank Dehne ● www.dehne.net “Good” Cases Frank Dehne ● www.dehne.net “Bad” Cases Frank Dehne ● www.dehne.net Experimental Verification Task: Design a protein that attaches to a yeast protein involved in DNA repair, thereby blocking its function. Target Yeast protein: YAL017W (PSK1) – DNA repair InSiPS generated protein: “Anti-PSK1”: HHHHHHSDNEHLHKCQRLKTRWKMARQFSDPQHNMYWIINWAQAM NIHADQNQEEEEELHDASVNNAEQYMAQCAPEEACQYPVRRSYGLH ATNCIERRKCCMIMYQHPTCRQWEAKNTCAISRAGKGVYWKGIIFMRA WKHWCTRRLVQ fitness: 0.465163 target score: 0.71832232 max non-target score: 0.35243136 (YLL039C) avg non-target score: 0.0720702297 Frank Dehne ● www.dehne.net Experimental Verification UV Light PSK1 DNA X mRNA mRN A Protein mRN Protein A mRNA mRN A Protein mRN Protein A Deletion DNA DNA Frank Dehne ● www.dehne.net X Anti-PSK1 mRNA mRN A Protein mRN Protein A Experimental Verification WT + Anti-PSK1 expressed PSK1 knockout Decreasing cell density WT WT (empty vector) Expression of Anti-Psk1 causes sensitivity to UV light. Equal numbers of cells serially diluted and exposed to 30s of UV light Frank Dehne ● www.dehne.net Future Projects Drug Design: Design synthetic proteins that attach to critical proteins in viruses, thereby inhibiting the virus. Possible Targets: - E-coli -… - HIV HIV (Capsid protein) Frank Dehne ● www.dehne.net
© Copyright 2026