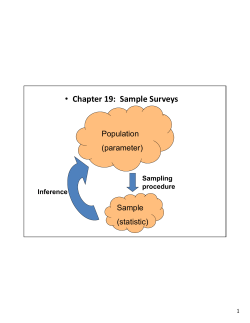
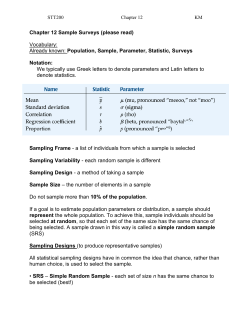
Appendix E Macroinvertebrate Sample Processing Error Report –
WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E Macroinvertebrate Sample Processing Error Report Appendix E Page 26 National River Health Program AusRivAS Quality Assurance and Quality Control Project Macroinvertebrate Sample Processing Error Report for The Department of the Environment and Heritage August 2004 Project Manager: Ross Bannister telephone +61 3 9550 1000 email [email protected] WATER ECOscience Pty Ltd ACN 064 477 989 Head Office 68 Ricketts Road Mt Waverley Victoria 3149 Australia Private Bag 1 Mt Waverley Victoria 3149 Australia telephone +61 3 9550 1000 facsimile +61 3 9543 7372 Wangaratta 1st floor NETC House 90-100 Ovens Street Wangaratta, Victoria, 3676 Carrum Eastern Treatment Plant Thompson Road Bangholme Victoria 3175 Gippsland 71a Argyle Street Traralgon Victoria 3844 Geelong 49 Carr Street Geelong, Victoria 3220 Hobart 20 St Johns Avenue New Town Tasmania 7008 Werribee Western Treatment Plant New Farm Road Werribee Victoria 3030 WATER ECOscience Report Number: 767 August 2004 Cover photo: RBA sweep sampling, Queensland Tasmania. © Commonwealth of Australia (courtesy of WATER ECOscience 2000) WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Foreword WATER ECOscience (formerly AWT Victoria) was engaged by the Department of the Environment and Heritage to undertake a study of Quality Assurance / Quality Control practices associated with AusRivAS data and collection methods. The study involved: • A national and international literature review of rapid biological assessment QA/QC techniques and procedures; • An independent external audit of the collection of habitat, physico-chemical and other related state / territory data used in the AusRivAS modelling and reporting processes; • an independent external audit of biological and other related state / territory data used in AusRivAS modelling and reporting processes, particularly for the development of AusRivAS models and the First National Assessment of River Health (FNARH), conducted under the National River Health Program (NRHP); and • an assessment of lead agency QA/QC procedures for data collection and data management. The macroinvertebrate sampling and processing component of the study was to assess and report on state agency performance for live-sort macroinvertebrate sampling, sample processing, and macroinvertebrate identification against existing criteria; as well as the incidence of data entry errors, and the effectiveness of existing QA/QC procedures. The audit covered data collected and sampling residues retained from randomly selected sites from the First National Assessment of River Health in each of the states / territories. Appendix E Page i WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Contents Foreword i 1 Introduction 1 1.1 Rapid Bioassessment 1 1.2 AusRivAS- National Context 2 1.3 Project Outline 3 1.4 Report Structure 4 2 Methods 5 2.1 Data Entry Audit 5 2.2 Live-sort Audit 6 Sample Collection Residue Processing Data manipulations WISE assessment 1. Live Sort / Whole Sample Estimate ratio 2. Bray-Curtis dissimilarity 2.3 QA/QC Procedures 3 Results 6 6 7 7 8 8 8 9 3.1 QA/QC procedures 9 3.2 Data Entry Audit 10 3.3 Live-sort Audit 10 4 Discussion 12 4.1 QA/QC Procedures 12 4.2 Data Entry Audit 12 Sampling records 4.3 Live-sort Audit 5 Conclusions and Recommendations 13 14 16 5.1 Key Findings and Conclusions 16 5.2 General Recommendations 16 6 References 19 Appendix E Page ii WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report List of Figures in Appendix E Figure 1 Location of the 24 sites assessed for the biological data entry audit and the 55 sites for the live-pick audit for Queensland in relation to all sites sampled during the MRHI program. 1 Figure 2 Location of the 52 sites assessed for the biological data entry audit and the 58 sites for the live-pick audit in New South Wales in relation to all sites sampled during the MRHI program. 2 Figure 3 Location of the 30 sites assessed for the biological data entry audit in the Australian Capital Territory in relation to all sites sampled during the MRHI program. 2 Figure 8 Location of the 28 sites assessed for the biological data entry audit for the Northern Territory in relation to all sites sampled during the MRHI program. 4 List of Tables Table 1 Table 2 Table 3 Number and proportion of macroinvertebrate identification sheets in each state / territory audited for data entry errors 5 Number of samples from each State audited for live-sort efficiency. 6 The lead agency of each state / territory audited and the relevant field sampling methods/guidelines employed by each (as was current during the field audit). 8 Table 4 Assessment of level of detail of field sheet and QA/QC procedures in audited states / territories. 9 Table 5 Errors associated with data entry of taxonomic information and abundances. 10 The number of samples and percentages of samples that failed QA/QC criteria for each State and habitat. 11 Table 6 Appendix E Page iii WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendices Appendix E:1 Sites selected for audits 1 Appendix E:2 Details of taxonomic data entry errors 5 Appendix E:3 Problems encountered in Data Entry Error and WISE Analysis 7 Appendix E:4 Whole Individual Sample Estimate (WISE) Analysis 9 Appendix E:5 Description of Habitats Sampled During State and Territory Field Audits. 11 Lead Agency Internal QA/QC Methods Live Picking Methods 20 35 Appendix E:6 Appendix E:7 Appendix E Page iv WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Summary This component of the AusRivAS Quality Assurance and Quality Control Project examined the performance of State and Territory lead agencies in performing livesort protocols and taxonomic data entry for AusRivAS models. The general findings were as follows: • documented QA/QC procedures are poorly developed and, although informal QA/QC procedures were often applied, they did not always show in agency performance. The level of QA/QC was difficult to assess as the level of documentation and sophistication varied across states and territories. Also, whilst some agencies had little or no documentation for QA/QC, their practices at the time of assessment may have included undocumented activities which conferred effective QA/QC on the data produced; • Approximately 75% of the live-sorted samples audited for this report passed the QA/QC criteria. This figure is a significant improvement on the first external QA/QC assessment of live-sorting procedures by Humphrey and Thurtell (1997) where only 47% of the samples passed the criteria used in the present report; • Approximately 30% of edge assessments and 21% of riffle habitat assessments using the current AusRivAS procedures may be erroneous. The remaining habitats assessed had approximately one tenth of samples with inadequate live-sorted fractions. The percentage of samples passing the QA/QC criteria in the present study are better than those reported by Humphrey and Thurtell (1997) who had up to 50% failure rates for edge samples and up to 31% of riffle habitats. It remains unclear why edge habitats had a greater failure rate than any other habitat type; and • basic record keeping, such as sample labelling and identification sheets, was found to be inadequate for some live-sort samples. The following suggestions are for the consideration of the Department of the Environment and Heritage and the state / territory agencies. General recommendations: • consistent documentation should be established for procedures associated with field and laboratory work, and QA/QC; • standard formats should be developed and applied for entry of field data, desk assessments, and laboratory work to ensure that all required data is obtained and calculated correctly; • a nationally agreed labelling standard should be developed to ensure that the labelling of samples is adequate for correct identification, processing, storage, and auditing; • nationally agreed QA/QC standards should be established embracing QA/QC processes as an integral component of AusRivAS assessment processes; provide guidance on field and laboratory procedures, equipment, sample Appendix E Page v WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report preservation for QA/QC assessment; target error rates for data entry and taxonomic misidentification; as well as live-sort performance; • the performance standards and targets should form part of coordinated national QA/QC program against which external auditing can be undertaken to review to assess its efficacy; and • QA/QC training, and assessment of operator competencies, should be undertaken in addition to existing AusRivAS training. Live-sort Procedures The substantial improvements in live-sort performance observed in this study underscore the value of external auditing, nevertheless some areas for improvement were highlighted, including: • improvements to live-sorting of edge samples; • extensive study of the relatively poor performance of the live-sort edge results in turbid habitats. Whilst it may be concluded that live-sort sampling bias is probably inherent to the method there is a need to better understand the biases and their quantify their implications for AusRivAS assessments. Appendix E Page vi WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 1 Introduction 1.1 Rapid Bioassessment Rapid biological assessment (ie. rapid bioassessment - RBA) can be used to describe two very different types of biological monitoring (Norris and Norris 1995). The first is a continual monitoring situation to detect trigger or alarm levels of organisms or toxicants. The second, and the subject of this review, refers to expeditious sampling of biota with rapid delivery of assessment results (Norris and Norris 1995). Benthic macroinvertebrate rapid bioassessment techniques and procedures were developed in 1977 in conjunction with the commencement of the RIVPACS program in October of the same year (Wright 1997, Davies 2001 pers comm). The United States Environmental Protection Authority further developed and expanded the RBA techniques to include fish and the work of Plafkin et al. (1989) was later used by several other countries (eg. Australia and Canada) to develop their own rapid bioassessment procedures (Norris and Norris 1995). Rapid bioassessment offers several advantages over the more traditional macroinvertebrate sampling methods, which involve a relatively large expenditure of time to collect, process and identify biological samples (Lenat & Eaton 1991). Rapid bioassessment reduces sampling effort, and therefore cost, by taking a relatively large sample instead of several individual replicates and reduces the number of organisms that must be processed by using a standardised subsampling procedure. In addition, rapid bioassessment programs often employ more efficient methods of data analysis than traditional biological assessment programs and produce results that are presented and summarised in a manner readily understood by non-specialists (Resh et al. 1995). Although rapid bioassessment of freshwater systems is now used in a number of different countries, only the United States, the United Kingdom, Canada and Australia conduct integrated, large-scale programs using comprehensive models that integrate macroinvertebrate and physico-chemical data to compare test sites to a benchmark or reference condition. The bioassessment programs used by these four countries vary in the extent to which they are applied and in their base method. However, all are based on similar theory and all require established and documented quality assurance and quality control systems to ensure that the integrity and veracity of the models used, and the results they produce, are maintained. Appendix E Page 1 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 1.2 AusRivAS- National Context The Australian River Assessment System (known as “AusRivAS”) was developed by the Cooperative Research Centre for Freshwater Ecology, in partnership with state and territory river management agencies, under the auspices of the National River Health Program (NRHP) funded by the Commonwealth Government. The NRHP was established in the Prime Minister’s Environment Statement in 1992 (O’Connor et al. 1996). The objectives of National River Health Program are to: • provide a sound information base on which to establish environmental flows; • undertake a comprehensive assessment of the health of inland waters, identify key areas for the maintenance of aquatic and riparian health and biodiversity, and identify stressed inland waters; • consolidate and apply techniques for improving the health of inland waters, particularly those identified as stressed; • develop community, industry and management expertise in sustainable water resources management and raise awareness of environmental health issues and the needs of our rivers. The NRHP, initially called the National River Processes and Management Program commenced in December 1992 (Davies 1994; O’Connor et al. 1996). The Monitoring River Health Initiative (MHRI) – a key component of the NRHP – used aquatic invertebrates to assess on a national level the ecological condition of Australian rivers (Smith & Kay 1998). As part of the MHRI more than 1500 reference sites were sampled across all states and territories during 1994/96 to establish the predictive AusRivAS models. The second phase of the NRHP utilised the AusRivAS models to undertake the First National Assessment of River Health (FNARH) (Smith & Kay 1998), later referred to as the Australia-wide Assessment of River Health (AWARH). The FNARH - AWARH commenced in 1997 and nearly 6000 sites have been assessed nation-wide. Australia is the first country in the world to undertake such a continental-scale assessment of the ecological health of its rivers (PIE 1998). The bioassessment component of AusRivAS uses a series of models to predict the composition of the aquatic macroinvertebrate community expected at a specific site in the absence of environmental stress (expected taxa (E)). This is compared with the macroinvertebrate community composition actually found at the site (observed (O)). AusRivAS assessments are reported as the ratio of observed to expected (O/E) taxa for the site, which are then assigned to a band indicating the extent to which a site has been impacted. Importantly, the veracity and national consistency of the AusRivAS–based river health assessments are reliant on the collection and entry of accurate and precise data. The current project, the National River Health Program - AusRivAS Quality Assurance and Quality Control Project, provides a national, external audit of data collected by the various state and territory government agencies using AusRivAS to assess river health in Australia. Information from this project augments previous quality assurance and quality control (QA/QC) work conducted under the NRHP Appendix E Page 2 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report and focuses on QA/QC during field operations, during subsequent laboratory sample processing and data entry, for both biological and environmental data associated with AusRivAS bioassessment for the NRHP. The audit encompasses all Australian states / territories and assesses their QA/QC procedures, field and analytical techniques and methodologies, personnel training, and data collection, validation and transcription. The purpose is to identify deficiencies and areas for improvement within state and territory agencies to ensure accuracy and consistency in the application of the AusRivAS model. WATER ECOscience (formerly AWT Victoria) was commissioned to undertake the National River Health Program - AusRivAS Quality Assurance and Quality Control Project. 1.3 Project Outline The National River Health Program - AusRivAS Quality Assurance and Quality Control Project was part of the Toolbox component of the Australia-Wide Assessment of River Health and involved a national, external audit of data collected by the various state and territory agencies using AusRivAS to assess river health. The objectives of the project were: 13. Assess and report on state agency performance for macroinvertebrate sampling, processing and identification procedures against existing criteria. 14. Develop criteria for assessing agency performance in the collection of environmental and habitat field data. 15. Assess and report on agency performance in the collection of environmental and habitat data and macroinvertebrate data. 16. Provide feedback and advice on the problems of staff performance in AusRivAS methods to state / territory agencies and to the new Training and Accreditation Project of AusRivAS. The project includes liaison with two other toolbox projects: • AusRivAS error analysis project - provide advice on actual error magnitudes in environmental data to enable evaluation of the consequences of errors associated with this type of data. • Training and Accreditation Project - liaison with the Principal Investigator to ensure deficiencies detected in the implementation of AusRivAS methods by lead agency staff can be addressed in any proposed training program. The AusRivAS Quality Assurance and Quality Control Project involves two types of audit, broadly categorised as: i) Veracity of macroinvertebrate sample processing and taxonomic identification, and ii) Collection and entry of field environmental (ie. habitat) data. This report deals with the first audit process and includes the macroinvertebrate sample processing, taxonomic identification, and entry of the biological data. In this audit an initial analysis of data entry errors was conducted for all states and territories, followed by an assessment of sorting accuracy for agencies us ing livesort procedures. Data entry errors were assessed by comparing the taxa and their abundance recorded on the macroinvertebrate identification sheets at the time of identification against that entered onto the agency databases. Appendix E Page 3 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report The accuracy of macroinvertebrate live-sort procedures was assessed for those states / territories using this method. These include Queensland, New South Wales, Tasmania, Victoria and Western Australia. The similarity of taxonomic composition from the live-sort fraction was compared with later identifications and counts on the residues retained from each sample. This comparison was performed using the Whole of Individual Sample Estimate procedure (WISE) (Humphrey & Thurtell 1997). The audit also included an assessment of quality assurance and quality control (QA/QC) procedures for the validation and transcription of biological data. 1.4 Report Structure This report is associated with objectives three and four of the project (see Section 1.3 above), and covers Phase 6: Assessment of Lead Agency Macroinvertebrate Sample Processing It includes auditing conducted over both the first and second stage of the project. The report reviews and reports on state / territory lead agency data entry for macroinvertebrate taxa and abundance, as well as live-sort performance. The effectiveness of QA/QC methods in controlling data entry errors and maintaining acceptable approaches for field collection of site environmental data, with particular emphasis given to data used as predictor variables in AusRivAS models. The report addresses the following three areas: 4. Audit of data entry errors associated with a random selection of sites sampled during the 1997 – 1999 FNARH program. Errors associated with missing or extra taxa and incorrect abundances were identified. 5. Audit of live-sort residues from the five States using live pick protocols. This included sub-sampling and picking of sample residues, and analysis of the performance of live-sorting using the WISE procedure. 6. Discussion of the impact of the various data entry error types and live-sorting performance on the outcome of AusRivAS river health assessments; and the adequacy of agency QA/QC practices. Possible improvements and further investigations are recommended. Appendix E Page 4 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 2 Methods The audit of state / territory lead agency macroinvertebrate sample processing commenced with an initial analysis of biological data entry errors. It was considered important that the data entry audit precede the live-sort audit to correct any data entry or taxonomic anomalies which may have confounded the outcome of the live-sort audit. The methods used by individual states and territories are listed in Table 16 below. More detail on the methods can be found in Appendix E:5 and E:6. 2.1 Data Entry Audit This audit assessed the accuracy with which lead agencies entered biological data from macroinvertebrate identification sheets into their database. This was performed by comparing the taxa and their abundance as recorded on the macroinvertebrate identification sheets (at the time of identification) against entries in the agency database. The lead agencies for each state / territory were requested to forward a list of all the sites sampled during the 1997 – 1999 FNARH program. From this list ten percent of sites were randomly selected for assessment. The locations for these sites are shown in the figures in Appendix E:1 together with all sites from the 1997 – 1999 FNARH program. The list of selected sites was then returned to the lead agencies with a further request for photocopies of the original macroinvertebrate identification sheets for these sites, together with an electronic copy of the data held in their database. Due to time constraints and the volume of data collected from each state / territory not all of the randomly selected sites were assessed. The number and proportion of data sheets audited for each state and territory are listed in Table 14. Table 14 Number and proportion of macroinvertebrate identification sheets in each state / territory audited for data entry errors Number of Audited Sheets Percentage of Audited Sheets Queensland 24 5% New South Wales 52 Australian Capital Territory Victoria State/Territory Number of Audited Sheets Percentage of Audited Sheets Tasmania 59 5% 5% South Australia 25 3% 30 5% Western Australia 27 7% 59 4% Northern Territory 28 6% State/Territory The electronic data was printed in a format to match the identification sheets. The paper copy of the data was marked with any errors or discrepancies found in comparison with the taxa and abundances recorded on the sheets. Data entry errors were assessed as follows: • Taxa missing TM) - taxa recorded on the identification sheet but not on the database. Appendix E Page 5 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Extra taxa (ET) – taxa recorded on the database but not on the identification sheet. • Incorrect abundance (IA) – incorrect abundance entered on the database, when compared to the identification sheet. It should be noted that there is a possibility of recording two errors (TM and ET) arising from one incorrect entry. For the case where one wrong taxa is entered there would be two errors generated, one corresponding to the extra taxa and the other to the missing taxa. To assess the situations where such error duplication may have occurred would require detailed examination of agency records and was therefore beyond the scope of this study. Differences between the identification sheets and the database were individually assessed to determine whether they represented genuine errors. Some discrepancies may have arisen from re-identification or QA/QC assessment by the lead agency and these were excluded from the analysis. 2.2 Live-sort Audit Sample Collection A total of 250 residue samples were assessed for the live-sort audit from the five States which conduct live picking (Table 15); the location of these sites are displayed in Appendix E:1 together with all sites from the 1997 – 1999 FNARH program. The collection of samples by each of these States is described in the manuals listed in Table 16, extracts of which can be found in Appendix E:7. Table 15 Number of samples from each State audited for live-sort efficiency. State Number of Audited Samples Percentage of Sites Audited Queensland 55 21% New South Wales 58 5% Victoria 51 12% Tasmania 59 12% Western Australia 27 5% Residue Processing The live-sort residues were sub-sampled in the laboratory at WATER ECOscience using equipment and procedures described by Marchant (1989). Subsampling involved placing the whole sample in a watertight box (35cm x 35cm), the bottom of which was divided into 100 cells. The box was sealed, gently shaken and then animals and other material removed from individual cells selected using random number tables. A total of 200 individual macroinvertebrates were subsampled in this manner according to the following criteria. The first 50 animals were identified, picked into vials and recorded. The following 150 animals were identified, but only new taxa not represented in the first 50 animals collected were picked and recorded. Finally, the remaining residue was placed in a large white sorting tray and scanned for 15 minutes with the aid of a magnification lamp. Any new taxa observed were picked Appendix E Page 6 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report and recorded. Recorded information includes a breakdown of what was collected during each component and the total overall percentage of residue that was subsampled. Pick results for each residue were later used to calculate the WSE for the sample from which the residue originated. Macroinvertebrate identification was to family level, except for Chironomidae (Diptera), which was identified to subfamily, and Collembola, Arachnida, and Oligochaeta which were identified to class or order level in accordance with accepted convention. Data was entered, validated and stored on the Zoological Database (MS Access) maintained by WATER ECOscience. Data manipulations The data was converted into the form necessary for input to the WISE database for assessment of the live-sort performance. Initially the data was checked to ensure that site, date and habitat details matched on both the Live Sort (LS) data and the Residue (RES) data for each sample. The WISE database will not run if there are any discrepancies in these details. Significant difficulties were encountered during the collation and manipulation of datasets sent to WATER ECOscience (see Appendix E:3). Generally these difficulties were associated with inadequate or erroneous detail on sample labels, identification sheets or database records; or taxa and site coding. All the data was then checked for duplicate records. The source of the duplication, identification sheets or the agency database or both, was then established and the appropriate corrections made. The WISE procedure was then run through one iteration to generate a printed output of the combined LS/RES data. These summary output tables were then assessed for taxonomic anomalies, generally coding errors, and appropriate corrections made to the database. As mentioned previously, most macroinvertebrate identifications were taken to family level and this is the taxonomic level at which data was inputted into the WISE database. However, if an animal was small or damaged and could not be identified to family level, the identification was taken as far as possible (usually order level). In the situation where a particular group of animals was represented in the dataset by both family level identifications and identifications at a lower taxonomic level (e.g. order), then all the animals within this group were combined back to the lowest common level. For example, if Zygoptera (unidentified) was present in either the LS or RES data then all families present within the order Zygoptera were summed under the heading of Zygoptera (unidentified). This was performed to avoid the possibility that taxa belonging to a representative of the family within the order (unidentified) was already represented in either of the datasets (i.e. a taxa would not be represented ‘twice’ in the dataset). The WISE procedure was then re-run through one hundred iterations. WISE assessment The WISE procedure uses two criteria to assess the performance of live-sorting the Live-Sort / Whole of Sample Estimate (LS/WSE) taxa number ratio and the Bray-Curtis dissimilarity measure (BC). These are described below and in more detail in Appendix E:4. Appendix E Page 7 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 1. Live Sort / Whole Sample Estimate ratio The LS/WSE ratio has been used previously to evaluate the performance of different sampling methods for the AusRivAS (Humphrey and Thurtell, 1997). The criterion for acceptable QA/QC of samples was that the number of taxa found in the LS must not be lower than 20% of those found in the WSE. That is, the LS/WSE must be greater than or equal to 0.8 for the number of live-sorted taxa to be considered representative of the whole sample. 2. Bray-Curtis dissimilarity Bray-Curtis (BC) dissimilarity values are derived by comparing the macroinvertebrate structure of each live-sorted sample with that remaining in the sample residue. Adjustment and normalisation of the dissimilarity values was performed as described in Appendix E:4. The BC value shows the similarity in the range of taxa collected during the live-sort compared to taxa present in the whole sample. The criterion for acceptable QA/QC performance based on presence-absence data is a maximum Bray-Curtis dissimilarity level of 0.35 (Humphrey and Thurtell, 1997). This criterion was used to assess the QA/QC of samples from the different States, values greater than 0.35 were taken to indicate live-sorted fractions unrepresentative of taxa in the whole sample. 2.3 QA/QC Procedures The methods (including QA/QC procedures) employed by each state / territory are summarised in Table 16, an assessment of the level of QA/QC is provided in Table 17 and further details can be found in Appendix E:6. Table 16 The lead agency of each state / territory audited and the relevant field sampling methods/guidelines employed by each (as was current during the field audit). State or Territory Lead Agency Relevant Field Sampling Methods/Guidelines Queensland: Department of Natural Resources & Mines Biological Monitoring and Assessment of Freshwaters using Macroinvertebrates, 1997 New South Wales: Environment Protection Authority NSW AusRivAS Sampling and Processing Manual, 2001 Australian Capital Territory: Environment ACT (CRC for Freshwater Ecology) ACT AusRivAS Sampling and Processing Manual, 2000 Victoria: Environment Protection Authority EPA Publication 604, 1998 Tasmania: Department of Primary Industry, Water and Environment Tasmanian AusRivAS Sampling and Processing Manual, 2000 South Australia: Environmental Protection Authority (Australian Water Quality Centre) SA AusRivAS Sampling and Processing Manual, 2000 Western Australia: Conservation and Land Management AusRivAS in Western Australia, 1998 Northern Territory: Department of Lands, Planning & Environment Northern Territory methodology, 2000 Appendix E Page 8 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 3 Results 3.1 QA/QC procedures The level of detail and thoroughness of QA/QC procedures varied between lead agencies. Table 17 provides an assessment of agency QA/QC based on both documented and accepted but undocumented practices. Most state / territory agencies were judged to have adequate to good procedures for animal collection, live-sorting and laboratory QA/QC procedures; NSW, Tasmania (excellent), Victoria, Queensland and Western Australia (good) had established procedures for data entry and validation. Sampling QA/QC procedures were mainly poor in all States and only adequate in Tasmania. Live sorting and laboratory QA/QC procedures were judged adequate to excellent. Three States, Victoria, Australian Capital Territory and South Australia, did not have documented QA/QC procedures for data entry or validation. However, in Victoria entered data is checked against data sheets a procedure which was not documented in the Victorian RBA manual. The documented data entry or validation procedures in the Northern Territory were poor and the remaining States had good or excellent procedures. Table 17 Assessment of level of detail of field sheet and QA/QC procedures in audited states / territories. QLD NSW 2 ACT VIC TAS3 SA WA NT 444 4444 4444 4 444 44 444 4444 – Sampling 4 4 4 4 44 4 4 4 – site assessment 44 444 44 4 444 4 4 4 – live-sorting procedure 44 X N/A 44 444 N/A 444 N/A 444 4444 4444 44 444 444 44 4444 444 4444 X 4441 4444 X 444 4 4 4 4 4 4 4 4 Documented Procedures Field sheet procedure (ie. Detailed instructions included with field sheets) Field QA/QC procedures Lab QA/QC procedures Data entry/validation – overall – independent check – range check5 4 4 Unless indicated otherwise ratings are based on QA/QC procedures documented in individual state / territory AusRivAS manuals. Level of detail was subjectively assessed by WATER ECOscience auditors and included supplementary information on accepted practices within agencies which are not formally documented. Number of ticks indicates thoroughness of documented procedures: 4 - poor, needs more detail; 44 - adequate; 444 - good; 4444 - excellent; X - indicates no established procedures; N/A – not applicable. 1 After data is entered it is checked against data sheets. However, this is not documented in the Victorian RBA manual. 2 Information based on Waddell (2001). QA/QC procedures are not documented in the NSW AusRivAS manual. Appendix E Page 9 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 3 Some QA/QC procedures are detailed in Tasmanian AusRivAS sampling manual, however, ratings given in table are primarily based on procedures documented in Krasnicki et al. 2001). 4 Based on supplementary information provided, procedures not documented. 5 Data entry/validation checks discussed in Section 4.1. 3.2 Data Entry Audit A total of 304 identification sheets and 6362 separate data entries were assessed for the three error types (Table 18). Overall, 11% of the identification sheets had errors and less than 1% of all data entered were incorrect. The most common error type was the incorrect abundance of animals in the sample being entered, nearly twice the rate of the other error types. Table 18 Errors associated with data entry of taxonomic information and abundances. State/Territory QLD NSW ACT VIC TAS SA WA NT Overall Number of taxa entries made 517 1086 553 1376 1212 739 393 486 6362 Number of id sheets assessed 24 52 30 59 59 25 27 28 304 Category Number of id sheets with errors 2 11 0 2 7 4 3 5 34 17 15 22 21 21 23 30 18 21 Taxa missing -electronic (TM) 0 6 0 0 3 0 3 2 14 Extra taxa (ET) 1 3 0 0 4 3 0 1 12 Incorrect abundance (IA) 1 4 0 2 3 1 1 10 22 Total entries made 0.4 1.2 0.0 0.1 0.8 0.5 1.0 2.7 0.6 Number of sheets 8.3 21.2 0.0 3.4 11.9 16.0 11.1 17.9 11.2 Average number of taxa per sheet Error types Error rates (%) The total number of entries and number of data sheets with errors varied between the states / territories. The Northern Territory had the greatest error rate, at 2.7% of all data entered, and the Australian Capital Territory the lowest with no data entry errors recorded. The greatest percentage of data sheets with errors was found in New South Wales with approximately one in five data sheets containing errors. 3.3 Live-sort Audit A total of 250 samples were assessed for QA/QC according to the L/W and BC criteria (Table 19). Of the 250 samples, 24% failed either the L/W criteria (less than 80% of taxa recorded in the LS compared with the WSE) or the BC criteria (at least 65% similarity between the two sample fractions) or both. The minimum failure rate for any habitat in any State was for Queensland riffles where 7% of samples failed. Appendix E Page 10 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Table 19 The number of samples and percentages of samples that failed QA/QC criteria for each State and habitat. Habitat State QLD NSW VIC TAS WA Data Bed Channel Edge Macrophyte Riffle Total Number of samples 14 27 14 55 L/W failed (%) BC failed (%) 7 14 11 15 0 7 7 13 At least one failure (%) 14 15 7 13 Number of samples 31 27 58 L/W failed (%) 45 41 43 BC failed (%) At least one failure (%) 16 45 0 41 9 43 Number of samples L/W failed (%) 30 20 21 14 51 18 BC failed (%) 20 5 14 At least one failure (%) 27 14 22 Number of samples 27 32 59 L/W failed (%) BC failed (%) 30 4 16 0 22 2 At least one failure (%) 30 16 22 Number of samples L/W failed (%) BC failed (%) At least one failure (%) Overall Number of samples L/W failed (%) 20 7 27 5 14 7 15 15 14 14 15 15 14 7 20 5 115 27 7 14 94 20 250 21 BC failed (%) 14 15 14 14 2 10 At least one failure (%) 14 15 30 14 21 24 Edge habitats had the greatest failure rate of the five types, with a 30% failure rate, followed by riffles (20%), channels (15%) and both macrophytes and bed habitats with 14% failure. The high failure rate of edge and riffle habitats appeared to be associated with samples from NSW where 45% and 41% failed respectively. However, the failure rate of edge samples was still nearly double that for riffles in Queensland, Victoria and Tasmania. Overall, the percentage of samples failing the L/W criteria was approximately double that of the percentage of samples failing the BC criteria and 7% failed both. The L/W criteria was failed more often than the BC criteria in New South Wales and Tasmania where the L/W failure rates were approximately 5 times and 10 times the BC rates respectively. The two criteria had similar failure rates for the other three States. Appendix E Page 11 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 4 Discussion 4.1 QA/QC Procedures The assessment of QA/QC procedures (Table 17) were based on both the documented procedures detailed in Appendix E:6 and supplementary information provided on undocumented standard practices within each agency. In assessing the level of QA/QC it must be recognised that, whilst some agencies had no documented procedures, their accepted practices encompass a high degree of QA/QC. Also, where there is low staff turnover and rigorous internal training a high level of QA/QC may be sustained without adequate documentation. Practices for data entry mostly included independent checking of entered data against the source documents. During the MRHI phase in NSW double entry of data was practiced, however in 1997 this was replaced by an independent check. In some States additional ‘range’ checks were also undertaken. These include identification and checking of outliers or data outside acceptable ranges. The criteria for outliers or acceptable ranges were obtained by plotting frequency histograms for the parameter of interest. Also, electronic data entry forms have been set up to mimic the layout of field and laboratory data sheets, and thereby minimise errors (NSW). 4.2 Data Entry Audit The overall percentage of data correctly entered from taxonomic identification sheets was high, with less than 1% of data entered with errors and approximately 90% of all data sheets with no errors. However, the percentage of sheets with data entry errors varied among states / territories with up to 20% of sheets with errors within one State. Data entry errors have the potential to affect the outcome of river health assessments using AusRivAS. Two of the three error types evaluated, errors associated with taxa on the identification sheet but not entered onto a database (TM error), and taxa entered onto the database but not on the identification sheet (ET error), have the greatest potential to affect river health assessment. Errors associated with the incorrect abundance being entered are unlikely to affect river health assessment as only presence/absence data is used in the AusRivAS predictive models. The total number of these two types of error (missing or extra taxa) that may affect river health assessment accounted for approximately half of all data-entry errors found, with up to 17% of data sheets in one State affected by these errors. This suggests that as many as 20% of site assessments may be affected by data entry problems within any one State. In addition, of all the data entered onto databases, only 0.4% (26 out of the 6362 entries made) were associated with TM and ET error types and generally only one taxon was affected per sample (see Appendix E:B), and two states / territories did not record any of these error types. An AusRivAS site assessment is based upon the ratio of animals found in a sample (Observed) to that predicted (Expected) to occur, the O/E ratio. Since about 20 taxa occur on average in samples, a TM or ET error, affecting only one taxon per Appendix E Page 12 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report identification sheet entry, may only increase or decrease an O/E ratio by 5%. This level of bias in the O/E ratio is unlikely to affect the health assessment of most streams or rivers. However, a single TM or ET data entry error may seriously affect the assessment of a site with a small number of predicted taxa. Using information from routine analytical laboratories, the acceptable rates of data entry or transcription error are probably in the range of 0.1%, or one error in 1000 entries. Using this rate as a target for AusRivAS QA/QC, nearly all of the states / territories exceeded this error rate. Also, the error rate did not appear to correlate with the degree of QA/QC practiced by each agency (Table 17). A number of practices can be implemented to reduce the rate of data entry errors, including: • independent checking of entered data against the source documents; • double entry of data and resolution of mismatches; • formatting data entry screens to mimic the layout of the field or data sheets; • range checks based on expected range for data (at a particular site); • use of optical character recognition (OCR) software in combination with redesign of the field or data sheets. Independent checking by another operator should be the minimum level of QA practiced. Double entry of data does not appear to offer significant benefits over and above independent checks and may be difficult to implement. The use of entry screens which match the data sheets should be achievable with current software packages. This is a basic measure which should be implemented to reduce error rates. OCR has been used successfully at NSW Fisheries for data entry however error rates are highly dependent on the design of the entry forms and the data being entered. The setup and operating costs may not justify the improvements in data accuracy at this stage of development of the technique. It is possible that future developments will make OCR more efficient and cost-effective. Other potential sources of error, such as the coding system used for macroinvertebrate families, may also make significant contributions to the overall error rates. Some coding systems are numeric or counter-intuitive and provide a greater opportunity for operators to enter or write the wrong taxa code. Adoption of a uniform national coding system utilising Vic EPA’s taxa codes (to species level, or to the level of identification) by all states and territories would minimise coding errors. Alternatively, consideration should be given to using family or species names rather than surrogate alpha-numeric codes. Sampling records A number of basic problems with data recording by agencies were encountering during the audit process. These included inadequate and ambiguous information on residue labels, identification sheets and database entries; as detailed in Appendix E:C. Because of these inadequacies, a number of residue samples were excluded from the live-sort audit. A matter of further concern is that the number of deficiencies appeared to be greater for the more experienced agency teams. Appendix E Page 13 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 4.3 Live-sort Audit Approximately 75% of the live-sorted samples audited for this report passed the QA/QC criteria. This figure is an improvement on the first external QA/QC assessment of live-sorting procedures by Humphrey and Thurtell (1997) where only 47% of the samples passed the criteria used in the present report. This suggests that improvements in AusRivAS procedures implemented after the first external audit have generally been successful and is an endorsement of agency efforts and the external audit process. The improvements undertaken following the 1997 audit included a minimum animal count of 200 or extended counting time, and the establishment of formal training facilities for AusRivAS assessments. However, the present level of live-sorted sample failures still means that as many as a quarter of all river health assessments using AusRivAS are potentially incorrect (generally by around +/-5% of overall condition) due to inadequate picking procedures generally by as much as 5% of assessed overall condition) due to inadequacies in picking or the live-pick protocol itself. The main habitats affected by inadequate picking in the present report were edge and riffle habitats with 30% and 21% of samples failing the QA/QC criteria. This suggests that approximately a third of edge assessments and one fifth of riffle habitat assessments using the current AusRivAS procedures may be erroneous. The remaining habitats assessed had approximately one tenth of samples with inadequate live-sorted fractions. The percentage of samples passing the QA/QC criteria in the present study are better than those reported by Humphrey and Thurtell (1997) who had up to 50% failure rates for edge samples and up to 31% of riffle habitats. It is unclear why edge habitats had a greater failure rate than any other habitat type but Humphrey and Thurtell (1997) suggested that edge habitats tended to have greater amounts of silt and detritus and therefore animals were harder to recover from the sample. However, the results of the present report and those of Humphrey and Thurtell (1997) suggest that significant improvements need to be made in the edge sample protocols and live-sort procedures. This may include washing silt from samples; or sorting through detritus by placing the whole sample in a bucket and sifting through smaller amounts at a time in the sorting tray; or by increasing the picking time. The percentage of samples failing the L/W criteria (21%) was approximately double the percentage of samples failing the BC criteria (10%). The failure of the L/W criteria has probably more serious implications for AusRivAS assessments than the failure of the BC values. This is because the O/E index uses the number of taxa collected in the live-sort in the numerator of the ratio, ie. the number of observed taxa. If the number of taxa in the live-sort was less than that collected in the whole sample the site would be assessed as in poorer condition that it actual was. These results suggest that as many as 20% of all sites, and 40% of cases in New South Wales, might be classified as in poorer condition than they really are. However the same biases may equally apply to the data for the reference sites. Therefore whether a different health assessment (poorer or better) would arise will depend on whether these biases may have moved one or other of the reference site or the actual test site out of band and in which direction. The net result is that there may not be significant changes to classifications. Humphrey and Thurtell (1997) indicated that small and cryptic taxa were commonly missed during the live-sorting process and suggested that extra training should be implemented to increase staff awareness of these taxa. In addition, changes to the live-sort protocol, namely the introduction of a target sample size of 200 animals rather than 30 minutes sampling, were implemented at this time (MRHI meeting Canberra, February 1997). A limited assessment of the data from this study Appendix E Page 14 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report suggests that there was no significant difference between taxa represented in livesort fractions versus the whole sample residue. It is not clear whether this indicates that the implementation of training, minimum counts and other improvements have been fully effective. Nevertheless, the extent to which small and cryptic taxa still contribute to the percentage of samples failing the L/W criteria should be the subject of on-going audit and assessment, to determine the need for further training and improvements in this area. Further, the Protocol Development Project (WATER ECOscience, 2003) found that whilst live-sort protocols were acceptable for clean sites, the presence of turbidity at some habitat sites led to poor live pick performance. Four protocol improvements were studied: 1. Use of visual aids during sorting; 2. Sorting fine and coarse fractions separately; 3. Use of a minimum time/number of animals; and 4. Selection of a minimum number of chironomids The study concluded that the proposed variants to the live-sorting protocol provided no increase in efficiency or veracity of results and that the standard method appeared as effective at retrieving an accurate representation of taxa at any one particular site. Furthermore, that poor live pick performance appeared to be related to a small number of animals in the live-sorted fraction and not the live-sort protocols themselves. Other factors, including lack of homogeneity between amalgamated samples and the efficacy of the live-sort protocols in some sample types, were also thought be contributors to poor live-sort performance. A more extensive study, involving a much larger number of sites and encompassing all live-pick Australian states and territories, was recommended to establish the validity and generality of turbid site live-sort QA/QC protocols. A more extensive study could include assessment of the types of taxa found in whole sample residues but not in the livesort fraction to ascertain if there is any bias for different habitat types in the different States. An alternative approach to this issue of probable bias, is to accept that live-sorting in the field and laboratory examination under a microscope are essentially two different methods which produce different types of data (Victorian EPA, 2001). Victoria considered that, like all sampling methods, there will be some inherent bias with livesorting with some taxa consistently over or under-estimated. The EPA concluded that samples should be collected in a consistent manner and the biases well understood. Appendix E Page 15 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 5 Conclusions and Recommendations The following key findings, conclusions and recommendations arise from this project, which assessed the QA/QC performance of live-sort protocols and taxonomic data entry. Whilst there are some deficiencies in data entry procedures noted, improvements in the live-sort QA/QC since the previous audit in 1997 are a positive finding. 5.1 Key Findings and Conclusions Documented QA/QC procedures are poorly developed and, although informal QA/QC procedures were often applied, they did not always show in agency performance; The level of QA/QC was difficult to assess as the level of documentation and sophistication varied across states and territories. Also, whilst some agencies had little or no documentation for QA/QC, their practices at the time of assessment may have included undocumented activities which conferred effective QA/QC on the data produced; Rates for data entry errors are higher than desirable, and vary significantly between agencies with an overall error rate of 0.6%. The most common error was incorrect entry of the abundance data, which does not influence the AusRivAS assessment. For the other error types (missing or extra taxa), since there are about 20 taxa per sample, a single such error would only increase or decrease the Observed / Expected animal ratio by 5%. This level of bias in an AusRivAS assessment is unlikely to affect the health assessment of most streams or rivers. Nevertheless, a single TM or ET data entry error may significantly affect the assessment of a site with a small number of predicted taxa. There appeared to be no correlation between the assessed level of QA/QC practices in each agency and the percentage of error rates. Inadequacies in basic record keeping, such as sample labelling and identification sheets, found during live-sort audits are unacceptable. Accordingly, the following recommendations are suggested for the consideration of the Department of the Environment and Heritage and the state / territory agencies. For completeness the recommendations include aspects of QA/QC not directly related to observations arising from the audit. 5.2 General Recommendations A QA/QC officer should be nominated for each state / territory agency, to coordinate all such activities and maintain consistency amongst operators, determine whether reproducible results are being obtained and performance target met. Documentation and data entry Procedures for field work, desk-based assessments and laboratory work should be adequately documented and made available for staff to follow in both the laboratory and the field. Standard forms should be developed for entry of field assessments and laboratory work to ensure that all required data is obtained and that calculations Appendix E Page 16 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report are carried out correctly. Similarly, nationally standardised labels should be used for field samples to ensure that they can be correctly identified and processed. All field samples should be either logged on field sheets or logged on return to the laboratory so that they can be tracked through processing and to provide an inventory of stored samples. Data entry and validation should be included in documented QA/QC procedures. These processes should include data validation by printing out database records for independent checking by another operator against the original data sheets. In addition, these documented procedures should include scanning for erroneous, extreme, outlier or ‘out of range’ values based on historical data or collective experience. Performance targets Targets for acceptable rates of error should be nominated immediately on an interim basis and adopted nationally as a matter of priority. Based on experiences gained in both this and the previous 1996 audit, the following are recommended as interim targets for adoption by state and territory lead agencies. They are not meant to be definitive but are designed to ensure adequate QA/QC targets and benchmarks are incorporated into AusRivAS processes while more considered standards are developed nationally. The objective of these interim targets is to improve the quality of AusRivAS outputs and provide end users of AusRivAS outputs with data of known levels of error. It is recommended that, at a minimum, state / territory agencies and those undertaking AusRivAS (such as the Murray-Darling Basin Commission and Natural Resource Management regional organisations) seek to achieve the following interim national QA/QC standards: 40. QA/QC processes are seen as an integral component of AusRivAS assessment processes and are be incorporated into AusRivAS protocols; 41. A suitably qualified and experienced QA/QC officer should be nominated for each state / territory agency to coordinate all such activities and maintain consistency amongst operators, determine whether reproducible results are being obtained, and performance target are being met; 42. Samples and accompanying field sheets from 10% of all AusRivAS sites being sampled should be preserved for QA/QC assessment (5% for internal QA/QC monitoring and 5% for external auditing). Where fewer than 10 sites are involved, samples from at least one site should be preserved; Performance targets should require that: 43. Fewer than 10% of field and taxonomic identification sheets have errors; 44. Less than 0.1% error rate for AusRivAS data transcription (data entered into state / territory data bases); 45. Less than 10% taxonomic misidentification at family level, and less than 10% taxonomic misidentification at species level; 46. Less than 20% of samples fail either LS/WSE or Bray-Curtis criteria; namely (a) (b) Appendix E LS/WSE ratio equal to or greater than 0.8, BC dissimilarity level equal to or less than 0.35. Page 17 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Also, QA/QC training should be undertaken, in addition to existing AusRivAS training, for all operators involved in field and laboratory procedures and data entry. This training should include ongoing assessment of competencies similar to that incorporated into the base AusRivAS training program. It is recommended that all such targets and procedures be integrated into a nationally coordinated QA/QC programs, and that this program then be reviewed to assess its efficacy through a process of external auditing. Live-sort procedures It is recommended that the external audit process undertaken in 1996 (Humphrey and Thurtell, 1997) be continued on a regular basis, to support an on-going improvement in the practices and QA/QC associated with AusRivAS assessments. The substantial improvements in live-sort performance since 1996 are validation of the external audit process, which includes feedback of results to participants and actions to improve performance. As edge samples were the main habitat type failing live-sort QA/QC significant improvements need to be made in the way that edge samples are live-sorted. A more extensive study is recommended to determine the poor performance of the live-sort protocol in particular turbid habitats. This study should include a larger number of sites for the States using live-sort protocols; and assess the types of taxa found in whole sample residues but not live-sort fractions to determine whether there is any bias for different habitat types in the different States. This more detailed study would provide an understanding of the biases present and whether their elimination or further reduction is warranted. It is probable that live-sort sampling bias is inherent to the method and that possible future improvements will not further reduce this bias. A recent assessment of potential improvements to live-sort protocols were judged to offer no improvement to the efficiency of the technique or the validity of the results obtained. Nevertheless, should this inherent bias be accepted, there is a need to better quantifying the livesorting bias and its implications for AusRivAS assessments. Appendix E Page 18 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report 6 References Davies, P. (1994) River Bioassessment Manual Version 1.0: National River Processes and Management Program, Monitoring River Health Initiative. LWRRDC, Canberra. Humphrey, C. and Thurtell, L. (1996) External QA/QC of MRHI agency subsampling and sorting procedures. Milestone Report to LWRRDC, December 1996. Lenat, D.R and Eaton, L.E. (1991) Comparison of a rapid bioassessment method with North Carolina’s macroinvertebrate collection method. Journal of the North American Benthological Society, 10: 335-338. Marchant, R. (1989). A subsampler for samples of benthic invertebrates. Bulletin of the Australian Society for Limnology 12: 49-52. Norris, R.H. and Norris, K.R. (1995). The need for biological assessment of water quality: Australian Perspective. Australian Journal of Ecology, 20: 1-6. O, Connor, N.A., Lloyd, L.N. and Moore, S.J. (1996). Evaluation of the National River Health Program. WATER ECOscience Report No. 56/96. PIE-Newsletter of Australia’s International and National Primary Industries R & D Organisations. International Acclaim for Aussie River Program August 98 edition. http://www.affa.gov.au/pie/98autumn/ Plafkin, J.L., Barbour, M.T., Porter, K.D., Gross, S.K. and Hughes, R.M. (1989). Rapid bioassessment protocols for use in streams and rivers. Benthic macroinvertebrates and fish, EPA/440/4-89/001, United States Environmental Protection Authority, Office of Water Regulations and Standards, Washington, DC. Resh, V.H., Norris, R.H. and Barbour, M.T. (1995). Design and implementation of rapid assessment approaches for water resource monitoring using benthic macroinvertebrates. Australian Journal of Ecology, 20: 108-121. Smith, M. and Kay, W. (1998). AusRivAS in Western Australia. An overview of the development and use of AusRivAS models for assessing river health in Western Australia. Department of Conservation and Land Management, WA Wildlife Research Centre, Wanneroo. Victorian EPA (2001). The Australia Wide Assessment of River Health; Final Report of the National River Health Program from Victoria. Environment Protection Authority, Victoria. Waddell, N. (2001) Australia Wide Assessment of River Health: New South Wales Program. Supporting Document to Final Report: Internal Quality Control and Quality Assurance Programs for the NRHP in NSW. New South Wales Environment Protection Authority. WATER ECOscience (2003). AusRivAS Protocol Development and Testing Project: Extended Analysis. WATER ECOscience Report No. 3055/03. Wright, J.F. (2000). An introduction to RIVPACS. In: Assessing the biological quality of fresh waters: RIVPACS and other techniques. Wright, J.F., Appendix E Page 19 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Sutcliffe, D.W. & Furse, M.T (Eds.) Freshwater Biological Association, UK. pp 1-24. Appendix E Page 20 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:1 Sites selected for audits The MRHI sites in each state / territory and the sites selected for auditing are shown in Figures 1-9. N MRHI Sites Audited Sites Figure 1 Location of the 24 sites assessed for the biological data entry audit and the 55 sites for the live-pick audit for Queensland in relation to all sites sampled during the MRHI program MRHI Sites Audited Sites Figure 2 Location of the 52 sites assessed for the biological data entry audit and the 58 sites for the live-pick audit in New South Wales in relation to all sites sampled during the MRHI program. Appendix E - appendices Page 1 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report MRHI Sites N Audited Sites Figure 3 Location of the 30 sites assessed for the biological data entry audit in the Australian Capital Territory in relation to all sites sampled during the MRHI program. MRHI Sites Audited Sites N Figure 4 Location of the 59 sites assessed for the biological data entry audit and the 59 sites for the live-pick audit for Victoria in relation to all sites sampled for the MRHI program. Appendix E - appendices Page 2 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report MRHI Sites N Audited Sites Figure 5 Location of the 59 sites assessed for the biological data entry audit and the 59 sites for the live-pick audit for Tasmania in relation to all sites sampled during the MRHI program. N MRHI Sites Audited Sites Figure 6 Location of the 25 sites assessed for the biological data entry audit for South Australia in relation to all sites sampled during the MRHI program. Appendix E - appendices Page 3 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report MRHI Sites N Audited Sites Figure 7 Location of the 27 sites assessed for the biological data entry audit and the 27 sites for the live-pick audit for Western Australia in relation to sites sampled for the MRHI program. N MRHI Sites Audited Sites Figure 8 Location of the 28 sites assessed for the biological data entry audit for the Northern Territory in relation to all sites sampled during the MRHI program. Appendix E - appendices Page 4 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:2 Details of entry errors taxonomic data Errors are presented as follows: Site X - Error type (number of errors): Taxa name (value entered)(correct value) Queensland Site 138110A - Incorrect abundance (1) - Caenidae (10 - 4) Site 136012A - Extra taxa (1) - Ochteridae Australian Capital Territory No errors New South Wales Abundances of over 26 entered as 99. Sphaeridae changed to Corbiculidae on database Site GWYD08 – Taxa missing (1): Chironomidae unidentified (49) Site MURR126 – Taxa missing (1): Daetidae (159)(99) Site RICH102 - Taxa missing (1): Psephenidae (6) Site RICH515 - Taxa missing (1): Corydalidae (2) Site LACH508 - Taxa missing (1): Diptera unidentified (1) Site LACH102 - Taxa missing (1): Telephlebiidae (1) Site TOWA570 - Incorrect abundance (1): Scirtidae (9 - 99) Site CLYD101 - Incorrect abundance (1): Hydropsychodidae (26 - 99) Site TURO602 - Incorrect abundance (1): Chironominae(32 - 5) Site BIDGM1 - Incorrect abundance (1): Gripopterigidae (5 -4) Site LACH115 - Extra taxa (3): Atyidae, Hydracarina, Parastacidae Victoria Site YY1 (15/12/98) - Incorrect abundance (1): Elmidae (A+(L))(14 - 15) Site EEX (09/04/97) - Incorrect abundance (1): Notonemouridae (5 - 4) Appendix E - appendices Page 5 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Tasmania Site E31 - Taxa missing (1): Tabanidae (2) Site FT23 - Taxa missing (1): Elmidae (1) Site G20 - Incorrect abundance (1): Parameletidae (6 - 5) Site GT18 - Incorrect abundance (1): Turbellaria (1 - 2) Site A15 - Incorrect abundance (1): Philorheithridae (7 - 8), Extra taxa (1): Odontoceridae (1) Site FT19 - Missing taxa (1): Philorheithridae (1), Extra taxa (1): Odontoceridae (1) possibly wrong taxa entered Site H10 - Extra taxa (2): Diamesinae, Helicophidae South Australia Site G106 - Incorrect abundance (1): Site J9 - Extra taxa (1): Cherax destructor (1) Site I51 - Extra taxa (1): Ranatra (1) Site I123 - Extra taxa (1): Ceratopogonidae pupae (1) Western Australia Site EWL01 - Incorrect abundance (1): Dytiscidae (78 - 76) Site HIL01 - Taxa missing (2): Diptera (unidentified)(1), Lepidoptera (1) Site SHA07 - Taxa missing (1): Trichoptera (unidentified)(1) Northern Territory Site AD08 (17/07/98) - Taxa missing (1): Lindeniidae (1) Site AD08 (16/10/98) - Incorrect abundance (4): Oligochaeta (46 - 54),Acarina (1 2), Tanypodinae (64 - 63), Chironominae (50 - 51) Site DA06 (31/10/98) - Taxa missing (1): Odonata (33) Site DA29 (2/11/98) - Incorrect abundance (1): Atyidae (8 - 7); Extra taxa (1): Syrphidae (1) Site DW22 (7/10/98) - Incorrect abundance (5): Nematoda (26 - 31), Planorbidae (1 - 2), Oligochaeta (23 - 21), Acarina (62 - 61), Tanypodinae (16 - 15) Appendix E - appendices Page 6 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:3 Problems encountered in Data Entry Error and WISE Analysis Considerable time was spent converting data supplied by lead agencies to a form suitable for the assessment of data entry errors and for import into the WISE database. The issues encountered included: • differing formats in which the lead agencies extracted the electronic data from their database; • errors or lack of site/date/habitat detail on sample labels, identification sheets and database entries; • identification of taxa to species level required re-coding and summing to family level; and the • use of taxa codes different from that used by WATER ECOscience (EPA Vic), necessitating re-coding. Queensland Considerable difficulties were encountered with compiling the comparable data sets. This was mostly due to the incorrect electronic data and missing identification sheets sent to WATER ECOscience. • Several residue labels were incomplete • Four samples were removed from the WISE analysis due to unresolvable uncertainties or incomplete data. New South Wales NSW had more problems with site data mismatches than any other State. The problems were associated with incorrect or incomplete site details entered on either the residue labels, identification sheets and/or the database. These were further confounded by the use of different site codes and identification sheets by the two agencies (EPA and DLWC) responsible for the sampling. • Two sites were removed from the WISE analysis due to unresolvable uncertainties or errors. Australian Capital Territory No problems with ACT data, not used in WISE analysis Victoria Considerable difficulties were encountered in determining the site locations of the residue samples supplied to WATER ECOscience due to poor sample labelling. • 13 residues did not have the site code written on the internal label (only a site description) and one residue did not have a habitat type/sample method. The Vic EPA residues internal labels had inadequate sample information. Eg. “King Parrot Creek - 9/4/97 - Kick Residue – Mike”. Appendix E - appendices Page 7 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Vic EPA electronic data had six date discrepancies when compared with ours, confirmed to be lead agency transcription errors. • Some of the Vic EPA data was identified to species level and some to family level. All species level identifications were converted to family level for WISE assessment. • The initial electronic data sent to WATER ECOscience was extracted in three separate sheets, this needed to be compiled and converted into a usable form for both the data entry error and WISE analysis. • During the data entry error assessment significant patterns of errors were encountered. The assessment was stopped and subsequent investigations revealed the lead agency had incorrectly extracted the data sent to WATER ECOscience. On delivery of the correct data the conversions and assessment were repeated. • Six samples were removed from the WISE analysis due to unresolvable uncertainties or incomplete data. Tasmania No problems with TAS data, no samples removed from the WISE analysis South Australia No problems with SA data, not used in WISE analysis Western Australia WA data had a different taxa coding system than that used at WATER ECOscience. Therefore, all taxa entries had to be re-coded before any analysis could be undertaken. • No discrepancies with WA data, therefore no samples removed from the WISE analysis Northern Territory No problems with NT data, not used in WISE analysis Appendix E - appendices Page 8 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:4 Whole Individual Sample Estimate (WISE) Analysis WISE: General overview and Bray-Curtis dissimilarity calculation The following is a very general overview of the Whole Individual Sample Estimate (WISE) procedure. For more detailed information refer to Mount and Humphrey (2001). The Whole of Individual Sample Estimate (WISE) procedure used in this study was based on methods initially described by Humphrey and Thurtell (1997) and later revised by Mount and Humphrey (2001). The WISE procedure has been incorporated in a Microsoft Access database developed for use with macroinvertebrate-based river health assessment under the AusRivAS scheme; it was designed to fulfil two major purposes: 1. Assess the performance of users in recovering small and/or cryptic taxa. 2. Allow a standardised assessment tool of operator performance which enables live-sort taxa recovery performance to be assessed for the whole NRHP. The WISE database compares the live-sorted component of a sample (LS) with an equivalent-sized component representative of the whole sample (prior to sorting) (WSE, or ‘Whole Sample Estimate’). The WISE database automatically calculates the Live-Sort/Whole of Sample Estimate (LS/WSE) taxa number ratio and various Bray-Curtis dissimilarity indices. The WSE is, in effect, an estimate of the taxa that would have been present in the original sample (Humphrey and Thurtell, 1997). The Bray-Curtis dissimilarity measure is calculated in WISE, for two samples, j and k, based on taxa 1 to N (indexed by i), as: ?∑ |xij - xik |?/ [ ∑ ? xij + xik? ] i i where xij is the abundance for taxon i in sample j (Mount and Humphrey, 2001). WISE calculates four different variants of the BC dissimilarity value for each sample: 47. Unadjusted BC dissimilarity for a sample size the same as the LS sample size. 48. Normalised unadjusted BC dissimilarity for situations where a sorted sample (ie the LS component) contains fewer than 100 individuals and the sample size is normalised to 100. 49. Adjusted BC dissimilarity where LS taxa that are unique to LS (ie. not found in the residue) are eliminated from the ensuing calculations (ie. prior to WSE calculation). 50. Normalised adjusted BC dissimilarity. Appendix E - appendices Page 9 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Adjusted BC values, involving the removal of taxa unique to the LS component, are provided because often live-sorting has removed all large, rare taxa and, consequently, such taxa would not be expected to occur in the residue and WSE. This report used adjusted BC dissimilarity values or, where the LS sample size was less than 75, normalised adjusted BC dissimilarity values. Appendix E - appendices Page 10 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:5 Description of Habitats Sampled During State and Territory Field Audits. The level of detail below is indicative of the detail in the manual, guideline or regulatory publication of each state or territory. Unless stated otherwise, descriptions of habitats sampled are derived directly from the AusRivAS methods manual for each state / territory and, where practical, text has been repeated verbatim. Queensland (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling//Qld/) Two habitat types are sampled, an edge sample and a bed sample. For bed samples the first choice is a riffle, however if this is absent samples should be taken from, in decreasing order of preference, a rocky bed or a sandy bed. Bed samples from the different habitat types are collected using slightly different techniques. Bed Samples Riffle habitat • Reach of relatively steep, shallow (<0.3 m), fast flowing (>2 m/s) and broken over stony beds. • Net size/type - standard 250 µm mesh, triangular dip net 250mm by 250 mm by 250 mm opening, 50-75 cm depth and with a 1 –1.5 m handle • Technique for sampling - while holding the net downstream with its mouth facing the sampling area, disturb the substratum by digging the foot well into the stones and turning them over. Turn and rub stones by hand to dislodge animals. Continue this process working upstream for 10m, covering both the fastest and slowest flowing sections. Stony/rocky bed habitat • Stony or rocky bed surface - if this is absent choose a silty area with plant litter or organic material • Net size/type - standard 250 µm mesh, triangular dip net 250mm by 250 mm by 250 mm opening, 50-75 cm depth and with a 1 –1.5 m handle • Technique for sampling - while holding the net downstream with its mouth facing the sampling area, disturb the substratum by digging the foot well into the stones and turning them over. If the rocks are too large to kick without damaging your foot, wash about 10 rocks of a range of sizes, scrubbing gently with the hands or a light brush into the net. Leave the rocks out of the water to allow cryptic specimens to emerge. These can then be hand picked. Appendix E - appendices Page 11 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Sandy/silty bed habitat • Sandy or silty bed with plant litter (not macrophytes) rather than an area of clean sand • Net size/type - standard 250 µm mesh, triangular dip net 250mm by 250 mm by 250 mm opening, 50-75 cm depth and with a 1 –1.5 m handle • Technique for sampling - sample a 10 m length of stream using a short sweeping action with the net whilst stirring up the bed with your foot. The suspended benthic animals are captured as the net sweeps through the cloud of suspended matter. Bed samples from Pool areas • Pools are zones of relatively deep, stationary or very slow flowing water over silty, sandy, stony or rocky beds. Water velocity is used to determine whether it is a pool or a run. • Net size/type - standard 250 µm mesh, triangular dip net 250mm by 250 mm by 250 mm opening, 50-75 cm depth and with a 1 –1.5 m handle. • Technique for sampling - Disturb the substratum by kicking with feet. If water is flowing, hold the net downstream with its mouth facing the sampling area. If there is no discharge, stir up the bed and use the net in a short sweeping action through the cloud of suspended matter to capture the suspended benthic organisms. Edge/Backwater Samples • Edges (or banks and underbank areas) occur along the bank where there is little or no current and extend to approximately 0.5 m from the bank. The area may be bare or have some terrestrial vegetation or tree roots. A backwater is a zone where the bank indents and forms a pool away from the main channel. backwaters may have a circular or back flow and a silty bed with accumulated plant litter and organic material. • Net size/type - standard 250 µm mesh, triangular dip net 250mm by 250 mm by 250 mm opening, 50-75 cm depth and with a 1 –1.5 m handle • Technique for sampling - using short upward sweeping movements at right angles to the bank, sample a total bank length of 10 m. Stir up the bottom while doing so, ensuring that benthic samples are suspended and then caught when sweeping through the cloud of suspended material. New South Wales (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/NSW/) For the purposes of AusRivAS a habitat is an instream environment within a sampling site that supports a distinct macroinvertebrate fauna. In NSW AusRivAS models were developed for Riffle and Edge habitats. These are defined below. Riffle habitat • The riffle habitat is an area of broken water with rapid current that has some cobble or boulder substratum. Appendix E - appendices Page 12 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Note: In cases where riffles do not have the type of substratum specified in this definition, the available habitat would generally be regarded as marginal or unsuitable. AusRivAS assessments from marginal and unsuitable habitats may be unreliable. For example, samples may be collected from broken waters with only pebble, gravel, sand or bedrock substratum or a combination of any of these. The results obtained from such riffles may be assessed as poor by AusRivAS even at relatively undisturbed sites. There is an advantage to this, however. In riffle zones where there is evidence to suggest an original cobble/boulder substratum has been covered by a pebble/gravel substratum as a result of human activities, AusRivAS assessments may give an indication of the taxa lost due to loss of instream habitat. In this case, the pre-disturbance substrata should be estimated and used for the assessment. Unreliable AusRivAS assessments may also occur as a result of difficult or dangerous sampling conditions. For example, sampling riffles where the substratum consists predominantly of large boulders may be difficult and potentially dangerous especially where flow is deep and fast. In addition, for situations where there are no riffles deeper than about 0.05m it may not be possible to obtain an adequate sample because many of the invertebrates released from the substratum when disturbed will not enter the net. This situation often coincides with slow flows and if riffle samples are collected under such circumstances, the results should be interpreted with this in mind. Macroinvertebrate data collected from small streams where the water velocity is very slow, even though broken water may exist, should also be treated with caution, as the riffle fauna in such streams may not be distinct from that found in the edge habitat. Natural riffle habitats are rare in Western NSW. Consequently the reference sites used to construct the riffle models in NSW are all from the eastern part of the State. Therefore using AusRivAS to assess unusual riffles on the Western Plains is not appropriate. • Net size/type - All macroinvertebrate sampling must be done with a kick net of 0.25 mm mesh size. The preferred net frame is one with a pentagonal shaped opening with a base of 35cm or greater. The net should be long enough to not cause backwash (60cm or more) and the net handle should be long enough (1.2m) to reach animals and microhabitats that are not immediately near the operator. • Technique for sampling - Locate the downstream end of the riffle zone within the site and begin sampling there. Disturb the substratum with your feet while holding the net downstream with its mouth facing upstream. Vigorously move the substratum about by digging your feet well into the cobbles and boulders. If necessary, turn and rub the boulders and cobbles by hand to dislodge organisms. Continue this process working upstream over a total distance of 10 metres comprising any number of discrete segments. Sampling should be conducted in both the fastest and slowest flowing sections of the riffle and at the maximum possible range of depths. It may be necessary to stop and rinse the net a couple of times during sampling to remove fine particles that can block the flow of water through the net which can cause backwash and loss of captured macroinvertebrates. It is also a good idea to thoroughly rinse the sample again once sampling is completed. This will assist in the sorting process by removing fine particles that can cloud water in the tray and obscure the animals present. Appendix E - appendices Page 13 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Edge habitat • The edge habitat is an area along the creek bank with little or no current. Suitable areas for sampling include an alcove or backwater with abundant benthic leaf-litter, fine organic/silt deposits, macrophyte beds, overhanging banks and areas with trailing bank vegetation. These areas are often indicated by the presence of surface-dwelling insects. Note: In lowland rivers the insides of sharp meander bends (point bars) and outside of sharp bends (steep eroded banks) should be avoided during sampling. A straight section is usually best, but suitable areas can sometimes be found just upstream or downstream of point bars (sandbanks). Ideally, sampling should be done at sections of the river that include an alcove/small bay between logs or red gums (on the lowlands of the Murray-Darling system) with logs and tree roots. Some river reaches have no areas along the edge that meet the requirement of having “little or no flow”. Such cases should be noted on the field data sheets and the results from the edge habitats at these sites treated with caution. • Net size/type - All macroinvertebrate sampling must be done with a kick net of 0.25 mm mesh size. The preferred net frame is one with a pentagonal shaped opening with a base of 35 cm or greater. The net should be long enough to not cause backwash (60 cm or more) and the net handle should be long enough (1.2 m) to reach animals and microhabitats that are not immediately near the operator. • Technique for sampling - Sweep the net over a total bank length of 10 metres comprising any number of discrete segments. Use sequential short sweeping movements at right angles to the bank (Figure 12). Stir up the bottom while doing so, such that benthic animals are suspended and then caught when sweeping through the cloud of suspended material. When sampling the edge habitat, try to sample as many different instream “structures” present in the reach as possible. Sweep the net in amongst tree roots, trailing bank vegetation, under overhanging banks and along logs if present. Do not, however, work into log crevices or use your hands or any means other than the net to extract animals. Macrophytes can be included in the edge habitat and should be sampled if abundant, however, small patches of macrophytes should not be deliberately sought for while sampling. Australian Capital Territory (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/ACT/) In the ACT, models have been created for both riffle and edge habitats. Areas of riffle and edge habitats that are representative of the reach should be chosen for sampling. The reach is defined as five times the mode bank-full width either side of the riffle sampling area, unless the bank-full width is less than 10m, then the minimum reach length is 100m (i.e., 50m either side of the riffle sampling site). Riffle habitat • The riffle habitat is one of flowing broken water over gravel, pebble, cobble or boulder, with a depth greater than 10 cm. • Net size/type - Samples are taken with a 250µm mesh rectangular or Dframed net with a 350mm wide aperture at the base. Appendix E - appendices Page 14 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Technique for sampling - Facing downstream, the operator should place the net directly on the substratum in front of the feet and vigorously disturb and dislodge the substratum by kicking and twisting the feet to a depth of approximately 10cm, slowly moving upstream employing this method. If fine particles block the flow of water through the net, stop and rinse the net before continuing. Separate lengths of riffle may be sampled if a continuous 10m section is not present. Record the collector’s name on the field sampling sheet along with the total length of riffle sampled, if less than 10m. Edge habitat • The edge habitat consists of slow flowing or still waters adjacent to the bank, preferably with overhanging or emergent vegetation, undercut banks, root mats or other suitable habitat providing cover and refuge for macroinvertebrates. • Net size/type - Samples are taken with a 250µm mesh rectangular or Dframed net with a 350mm wide aperture at the base. • Technique for sampling - macroinvertebrates are collected by vigorously sweeping from a distance of approximately one metre from the bank to the bank edge, disturbing the emergent and overhanging vegetation in the water if present. The operator should slowly move upstream for a distance of 10 metres employing this method. Separate lengths of edge may be sampled if a continuous 10m section is not present. Record the collector’s name on the field sampling sheet along with the total length of edge sampled, if less than 10m. Victoria (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/Vic/) Riffle habitat • Typically rocky riffles where the flow is rapid and turbulent, but gravel or sand bars can also be sampled as long as there is flow over the substrate • Net size/type - opening 30 by 30 cm, mesh size 250 µm net length approximately 1 m • Technique for sampling - sampler disturbs the streambed by vigorously kicking while holding a collecting net down current, debris dislodged by the kicking is collected in the net as the sampler moves upstream, continually kicking the streambed for 10 metres, typically taking 5 to 10 minutes Edge habitat • Areas of little or no current along the edge of the stream – around overhanging vegetation, snags and logs in backwaters and through beds of macrophytes • Net size/type - opening 30 by 30 cm, mesh size 250 um, net length approximately 40 cm • Technique for sampling - net is swept along the edge of the stream in areas of little or no current and around overhanging vegetation, logs and snags in backwaters and through macrophyte beds for 10 metres, typically taking 5 to 10 minutes Appendix E - appendices Page 15 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Tasmania (AusRivAS Manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/Tas/) In Tasmania, models have been created for both Riffle and Edge habitats. The reach is defined as 100m of stream length and within the stream only. Areas of riffle and edge habitats that are representative of the reach should be chosen for sampling. Riffle habitat • The riffle habitat is one of flowing broken water over gravel, pebble, cobble or boulder, with a depth greater than 10 cm. • Net size/type - samples are taken with a 250 mm mesh kick net with a 280x340 mm opening. • Technique for sampling - facing downstream, the operator should place the net directly on the substratum in front of the feet and vigorously disturb and dislodge the substratum by kicking and twisting the feet to a depth of approximately 10 cm, slowly moving upstream employing this method. Every 2-3 metres the net should be rinsed to remove fine particles which may be blocking the flow of water through the net. Separate lengths of riffle may be sampled if a continuous 10 metre section is not present. Note total length of riffle sampled if less than 10 metres. Edge habitat • Consists of slow flowing or still waters adjacent to the bank, preferably with overhanging or emergent vegetation, undercut banks, root mats or other suitable habitat providing cover and refuge for macroinvertebrates. • Net size/type - samples are taken with a 250 mm mesh kick net with a 280x340 mm opening. • Technique for sampling - macroinvertebrates are collected by vigorously sweeping from a distance of approximately one metre from the bank to the bank edge, disturbing the emergent and overhanging vegetation in the water if present. The operator should slowly move upstream for a distance of 10 metres employing this method. Separate lengths of edge habitat may be sampled if a continuous 10 metre section is not present. Note total length of edge sampled if less than 10 metres. The net should be thoroughly rinsed to remove silt, mud, and fine detritus. South Australia (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/SA/ ) Areas with riffle and edge habitats that are representative of the conditions found within the reach of interest along a creek or river should be chosen for sampling. The site is defined as a 100m section of the stream as stipulated in the bioassessment manual (Anon 1994). In South Australia, models have been created for both Riffle and Edge habitats. Appendix E - appendices Page 16 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Riffle habitat • Areas of shallow turbulent water flowing over a substrate, which is usually cobble, pebble and/or gravel, but may include sand, detritus, roots, etc. • Net size/type - macroinvertebrate samples were taken using a triangular framed 250 mm mesh pond net (35 x 30 x30cm). • Technique for sampling - The substrate was disturbed by vigorous kicking and rocks were rubbed by hand to dislodge organisms into the net that was held immediately downstream. Note that sampling proceeded upstream or was conducted in a manner that prevented the contamination of samples due to drift and disturbance while working the site. The sampled area was not necessarily contiguous, and attempted to encompass all the microhabitats available. Edge habitat • Areas of little to no current, or aquatic vegetation, often in quite deep water. It may have overhanging or emergent vegetation, undercut banks, root mats or other suitable habitat providing cover and refuge for macroinvertebrates. • Net size/type - macroinvertebrate samples were taken using a triangular framed 250 mm mesh pond net (35 x 30 x30cm) • Technique for sampling - The net was moved in sweeping actions through the water column as the sampler moved along the bank, and the sediment was kicked to ensure benthic organisms were collected in the sample. Three rocks where also rubbed by hand (when present) and the dislodged acroinvertebrates included in the sample. Note that sampling proceeded upstream or was conducted in a manner that prevented the contamination of samples due to drift and disturbance while working the site. The sampled area was not necessarily contiguous, and attempted to encompass all the microhabitats available. Western Australia (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/ Man/Sampling/WA/) Riffle habitat • Areas of steep broken water with rapid current and stony substrate • Net size/type - 350 by 250mm opening, 250 µm mesh, 50-75 cm depth and 11.5 m handle • Sampling technique - disturb the substratum while holding the net downstream with its mouth facing the disturbed area. Kick the substratum by digging the foot well into the stones and turning them over – turn and rub stones by hand to dislodge animals. Continue this process working upstream over a distance of 10 metres, covering both the fastest and slowest areas of the riffle. Do not include material from macrophytes or woody debris located in the riffle. Macrophyte habitat • An area of dense aquatic vegetation Appendix E - appendices Page 17 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Net size/type - 350 by 250mm opening, 250 µm mesh, 50-75 cm depth and 11.5 m handle • Technique for sampling - vigorously sweep the net within the aquatic vegetation over a length of about 10 m aim to sample the upper middle and lower portions of the plants. A combination of short lateral sweeps with vertical lifts will aid in dislodging and catching suspended animals Channel • The central part and margins of the main channel of a stream in areas without riffles, macrophytes or pool rocks • Net size/type - 350 by 250mm opening, 250 um mesh, 50-75 cm depth and 11.5 m handle • Sampling technique - vigorously sweep the net through the water column using short vertical lifts to disturb the substrate and catch the suspended organisms. Continue this process along the channel for a distance of about 10 m. Pool rock habitat • A pool with little or no current and large numbers of rocks • Net size/type - 350 by 250mm opening, 250 µm mesh, 50-75 cm depth and 11.5 m handle • Sampling technique - remove about 10 – 20 rocks of a range of sizes from the stream into a bucket and rub the rocks by hand to dislodge the animals Northern Territory (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/NT/) Edge habitat • Ideally a vertical or sloping section of the riverbank containing abundant root matter which is immediately adjacent to a pool. Areas to avoid when sampling edge habitats are: pandanus roots, adjacent macrophytes (water plants) and large undercut banks. Pools can be of variable depth from 30cm to 3cm or more. • Net size/type - triangular framed net with 350 mm wide aperture at base, 250 µm mesh, long handled also used is a three pronged rake device used by a second operator to disturb the sediments and vegetation in front of the net • Sampling technique - macroinvertebrates are collected by the rake operator agitating the root matter in the water and adjacent to the bank. The net operator sweeps vigorously back and forth following the rake operator and capturing the material dislodged into the water column. In still waters the net is kept mobile to ensure water current is generated through the net and the collected macroinvertebrates do not swim out of the net. Both operators move upstream for a distance of 10 m. Appendix E - appendices Page 18 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Sand/Silt bed habitat • Area of uniformly deposited sand or silt. Generally the surface is horizontal and at a depth of no greater than 40cm. Areas of thick detritus (organic matter eg. .leaves) and algae are avoided. • Net size/type - triangular framed net with 350 mm wide aperture at base, 250 µm mesh, long handled also used is a three pronged rake device used by a second operator to disturb the sediments and vegetation in front of the net • Sampling technique - macroinvertebrates are collected by the rake operator facing the water and raking in 30 cm scratches across the sand/silt substrate. The raking pattern is achieved by extending the rake out in front and then raking towards the body. The net operator faces the water also and employs a ‘figure of eight’ sweeping motion which collects the dislodged material. The net operator should endeavour to keep the net sweeping the habitat surface without collecting large amounts of sediment. Both operators move upstream for a distance of 10 m. Appendix E - appendices Page 19 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:6 Lead Agency Methods Internal QA/QC The internal QA/QC protocols varied between States and this was reflected in each agencies method manual. The level of detail below is indicative of the detail in the manual, guideline or regulatory publication of each state or territory. Where practical, text has been repeated verbatim from the state / territory AusRivAS methods manual. Queensland (AusRivAS manual:http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/Qld/) Field The residues of ten percent of all samples taken in the field are retained for analysis. Half of these samples are put aside for external analysis; the other half are subsampled and 10% of each sample is analysed by the unit’s staff. The data is analysed, compared to the sample picked in the field, reports written, and presented as part of the milestone reports. Laboratory • As the samples arrive in the laboratory from the field, an audit is taken. The site name and type of habitat sampled is recorded. This list is compared against the field sheets when they arrive to ensure that no samples have been lost. The vials are inspected for cracks etc which might allow the sample to dry out. Low alcohol levels are topped up and the vials stored upright according to sampling run and district from which they are collected. The QA/QC residue samples are audited also and checked against the list of requested samples. The samples are stored in sealed polydrums until required. • As the paperwork arrives in the laboratory from the field, it is stored in ring binders according to sampling run and district. The field sheets are checked by the staff member that assisted in the field in each district. Any empty fields are completed by either the biologist that attended the collection, or the district staff that undertook the sampling. The photos are stored with the field sheets as are the major ion and nutrient water quality results from GCL. Taxonomy • Internal QA/QC checks are performed on staff, by staff, on a regular basis. At each round of QA/QC, a person is assigned to analyse a sample identified by another. Samples identified during the previous fortnight are selected at random and re-identified. The resultant taxa lists are compared and discrepancies in identification checked by other staff in the unit. Any errors are discussed with the original identifier (both misidentifications and errors of enumeration) and a report prepared which is read and signed by all members that underwent the QA/QC check. Appendix E - appendices Page 20 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Data Entry • Taxonomic data is entered electronically (EXCEL database) as the samples are completed. At the completion of a district, the electronic database is verified against the data sheets. This is a 2-person task, where one person reads aloud the entries from the data sheet and the other person checks these results against the electronic database. Corrections are made to the database as the verification proceeds. When a full run is completed and fully verified, the data is transferred electronically to an ACCESS database. Random checks are implemented to ensure that the data transferred successfully. • Physical and chemical data is entered directly from the field and GCL sheets onto an Excel database. Parameters that are measured both in the field and as part of the GCL analysis (Conductivity, pH, turbidity and alkalinity) are compared and, if the results are compatible, the field results are recorded on the database. If there is a major discrepancy, the cause is investigated by referring to other sampling occasions. A picture of the site is built up and the more appropriate readings are used in the database. Substrate composition is checked by adding the components and ensuring all samples measure 100%. Formulae have also been used in the database to calculate mean phi of the substrate and to rate chemical measurements. On completion of data extraction to Excel, random checks are done to ensure that data was transcribed correctly. The data is than transferred electronically to the ACCESS database. Random checks are implemented to ensure that the data transferred successfully. • Several descriptive variables are also extracted from the field sheets to describe things such as canopy cover, bank stability, channel condition. These ratings are entered directly to an Excel spreadsheet. On completion of the data extraction, the database is transferred electronically to the ACCESS database. Random checks are implemented to ensure that the data transferred successfully. • Once the data is stored in ACCESS, each parameter is checked for outliers. These are referred back to the field and laboratory sheets and corrected, if necessary. Mapping • Several parameters are extracted from maps. Most are done by the district hydrographers, checked by other hydrographers and sent to us electronically. We rely upon the data being checked prior being sent. Latitude and longitude values are mapped to ensure that the sites map to the correct position. • Data is also extracted from electronic databases such as BOM, DRF and vegetation overlays. These data are entered directly into an Excel spreadsheet and later matched to the appropriate sampling site. This data is then transferred electronically to the ACCESS database. Random checks are implemented to ensure that the data was entered correctly and was transferred successfully. New South Wales (Waddell 2001) Note: No details of New South Wales QA/QC procedures are provided in the NSW AusRivAS Sampling and Processing Manual. In reference to such procedures the Appendix E - appendices Page 21 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report manual merely indicates that ‘It is also important that in undertaking sampling for AUSRIVAS, particularly when large-scale monitoring programs are concerned, appropriate quality control and quality assurance procedures are followed. Details of quality control and quality assurance programs followed during the MRHI and AWARH in NSW can be found in Waddell (2001).’ Quality control and assurance programs for the NRHP were undertaken both at the national level and within each state and territory Program. During MRHI QA/QC programs were initiated at the national level to address issues of sorting and macroinvertebrate identification. A second national QA/QC program was also recently implemented to deal with sorting issues for the AWARH phase. The individual states and territories assumed responsibility for QA/QC for identifications during AWARH. The information presented below outlines the QA/QC programs undertaken out at the state / territory level for NSW. Field measurements • Water quality meters were calibrated prior to each sampling event and checked daily during sampling. • Alkalinity was measured both in the field and in the laboratory from frozen water samples. Values were compared and suspect data flagged and excluded from analyses. • Field measurements such as stream width and riffle depth were regularly confirmed using a measuring tape and more subjective measurements such as disturbance rankings were regularly compared between team leaders to ensure consistency between sampling teams. • Site attributes such as site code, name, position and elevation were checked using topographic maps and/or GPS on each sampling occasion. Positional accuracy was reconfirmed in the office using GIS. Data entry and storage • All data collected during the NRHP for NSW were entered and stored in an Oracle database. To ensure completeness of records in the database, all samples collected in the field were entered into a field master form within the database. This was done immediately following all sampling trips and included information such as site code, date and the habitats from which biological samples were collected. A complete record of all samples collected was therefore readily available. • To help minimise errors associated with data entry, electronic data entry forms were set up to mimic the layout of the field and laboratory datasheets. • Range checks were also in-built to highlight unusual or incorrect values for given variables (such as a pH value >10). • All entered data was then checked. During the MRHI phase this entailed the double entry of all field and biological data into a QA/QC table followed by electronic comparison and subsequent checking of inconsistent results. In 1997 this procedure was replaced by a separate visual check of inconsistent data between hardcopy records and database records by another operator. Errors were then rectified and changes noted on the original datasheet. To provide a measure of NSW EPA QA/QC Document accuracy in data entry, a second checking procedure was undertaken on the field data from 25 Appendix E - appendices Page 22 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report randomly selected samples in each season. All errors were recorded and these data were analysed to provide an assessment data entry accuracy. Macroinvertebrate identification • Five percent of samples were selected from each season on a stratified/random basis ensuring all identifiers were considered and a range of habitats and biogeographic regions represented. Selected samples were then re-identified by an experienced staff member according to the guidelines presented in Hawking and O’Connor (1997a). • Error rates including Percentage of New Taxa and the Bray Curtis Dissimilarity Index were then calculated for each of these samples as specified in Hawking and O’Connor (1997b). • In addition, Sorrenson’s index (Bennison et al., 1989) was also calculated to provide an alternative estimate of dissimilarity between original and QA/QC samples. This index was considered more appropriate than Bray Curtis for reflecting errors that may affect AUSRIVAS results, which are based on presence/absence data, as it uses total taxa numbers and not relative abundance. • In accordance with the MRHI QA/QC program (Hawking and O’Connor, 1997c) < 10% error was deemed acceptable and therefore samples with a ‘new taxa’ percentage of 10% or greater and/or a Sorrenson’s index of less than 0.91 failed the QA/QC test. • All identification errors were compiled and appropriate follow-up action implemented to rectify mistakes and improve identification performance. Follow-up action was also undertaken to address identification problems highlighted in the national QA/QC program conducted on samples collected during the MRHI phase (Hawking and O’Connor, 1997c). Biological Data Screening To ensure that only quality-assured samples were included in model development and performance testing, a formal procedure was undertaken to screen all biological data collected at reference sites. A set of criteria was developed and biological samples that failed to pass the quality control and assurance procedure were flagged in the database and excluded from use in all modeling procedures. Biological samples were assigned a ‘fail’ for quality control and assurance if they contained: • An unusually low number of taxa for the site (as compared to other samples collected at the site for the same habitat and season) and/or, • Unusually low O/E results and a different fauna composition than expected for the site in the relevant season and habitat (as compared to other samples collected at the site) and/or, • A lower than expected number of cryptic taxa (as compared to other samples collected at the site and similar sites for the same habitat and season). And satisfied one or more of the following criteria: • Sorted by an untrained or inexperienced operator, • Collected by an untrained or inexperienced operator, Appendix E - appendices Page 23 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Collected from a marginal habitat eg. small bedrock riffle or a fast flowing edge, • Collected from an unusual site eg. acid stream, • Sorted under low or artificial light conditions eg. dusk or motel room, • Sampled with limited access to available habitats eg. steep, slippery banks, deep pools or very fast flowing riffles, • Sampled under extreme circumstances eg. heavy rain, • Sampled under extreme flow conditions ie. during or immediately following a flood or drought, • Collected outside the acceptable date range for a given season. This screening procedure was also applied to some test site data where replicate samples were available for comparison. The quality of test data, however, is a lot harder to assess than reference data. Whereas data collected in other years/seasons from the same site and in the same year from similar sites could be used as a benchmark for assessing reference site data, such benchmarks are not available for test sites because it is neither possible nor appropriate to attempt to anticipate results from test sites. Even when replicates were available from a disturbed test site it was often not safe to assume that there should be consistency in the results over time because in most cases the degree of disturbance would change greatly over time. Consequently the screening procedure for the test site data was probably less reliable and depended on the results of the reference data screening procedure to identify possible problems such as consistently poor sorters, flood or drought extremity, difficult sampling conditions etc. Environmental Data Screening Map/GIS derived data All predictor variables derived by topographic maps or GIS including elevation, distance from source, latitude, longitude, slope and mean annual rainfall, were checked and screened for unexpected results. Field data A rigorous screening procedure was undertaken to ensure only reliable field recordings were used in all aspects of model development, testing and site assessment. This followed the finding of a recent internal study, conducted to assess temporal variation in AUSRIVAS outputs, that one of the major factors affecting group probabilities and hence O/E values between samples from the same reference site was variation in environmental data. This was particularly evident for substrate variables such as clay and silt where large differences were evident between different sampling occasions. As a consequence, potential predictor variables including substrate composition, mode stream width, mode riffle depth and field alkalinity readings were screened for all reference and test samples. Records that were inconsistent with other samples and/or inconsistent with the checkers knowledge of the site were recorded as unreliable and eliminated from any analysis. For modeling purposes substitute values were then derived for all deleted and previously missing environmental records. The mean value of all quality assured records for the site was used for this purpose. If no quality assured values were Appendix E - appendices Page 24 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report available for a site an estimate was derived from values recorded at similar sites in combination with the samplers knowledge of the site. For alkalinity, field records were also compared to values derived in the laboratory from frozen water samples. If consistent with laboratory-derived values field records were used preferentially for modelling purposes. If reliable field data were not available for a sample the lab value was used followed by the mean value of all quality assured field records. Australian Capital Territory (AusRivAS manual http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/ACT/) Field • It is critical that once the field sampling sheet has been completed, it is rechecked to ensure no measurements have been missed. The discovery of missing data once back in the laboratory can mean returning to the site, a costly mistake in both time and money, and one that could have been easily avoided. It is also critical that measurements of zero are recorded as such on the field sampling sheet and not left blank. A blank value may be interpreted as missing data when being entered into a database by another person and can result in the site not being assessed. Laboratory Quality control/quality assurance (QA/QC) procedures are designed to establish an acceptable standard of macroinvertebrate sorting and identification. The quality control component reduces the level of error in sorting and identification, while the quality assurance component provides potential users with the assurance that the accuracy of results is within controlled and acceptable limits. Sorting • For new persons, projects, or sampling runs, quality control staff should check the residues of the first five samples sorted for missed organisms. In order to pass the QA/QC process, = 95% of the total number of organisms in the sub-sample must be recovered. If one of the first five samples fails the QA/QC protocol, the process begins again until five consecutive samples have passed. • Following the initial five samples, a random selection of two samples in the following ten, two samples in the following 30 and two samples in the following 50, will be checked. If any of these samples fail the QA/QC protocol, the person must again pass five consecutive samples. • Staff checking samples will have adequate experience in sorting. Where possible, QA/QC of sample residues should be conducted by more than one person to avoid bias and increased workloads. If less than 95% of the organisms are recovered from the sub-sample, commonly missed taxa should be shown to the person and suitable instruction given to rectify the problem. Laboratory sample record sheets are to be fully completed by the person conducting the QA/QC check. Error/action codes are to be recorded on laboratory sample record sheets if appropriate. Appendix E - appendices Page 25 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report QA/QC error and action codes Error codes • CC Number of organisms recovered from the sub-sample represents less than 95% of the total number of organisms in the sub-sample. • IE Identification error (i.e., “Percent Taxa Error” and/or the “Percent Incorrect Identifications”) greater than 5%. • LE Labelling error • SE Sub-sampling error – e.g., if the sample was stored in more than one sample container and only one container was sorted. • WE Washing error – some sample bypassed sieve when rinsing • DE Data entry error on data sheet • CE Calculation error – mathematical error on data sheets Action codes • LC Labels corrected – contact person who collected the sample if error is on the original sample label • SC Sample re-subsampled, processed, re-checked and data sheets corrected • WC Material bypassing the sieve caught in washbasin, sample combined and rewashed • WI Material bypassing sieve lost, partial sample processed • DC Data entry corrected (strike out incorrect entry with one line and write in the correct entry, initial). Identification Two methods are used to calculate a sample’s identification QA/QC result. These are the “Percent Taxa Error” and the “Percent Incorrect Identifications”. A sample must pass both methods to achieve an overall pass. Percent Taxa Error A "Taxa Error" occurs when a mis-identification results in the loss or addition of a taxon. The “Percent Taxa Error” is the "Number of Taxa Errors" divided by the "Total Number of Original Taxa", multiplied by one hundred. Samples pass if the "Percent Taxa Error" is less than or equal to 5% at the family level (10% at species level). The manual provides examples (not included here) of how to calculate ‘Taxa Error’. Percent Incorrect Identifications The “Percent Incorrect Identifications” is the "Number of Organisms Incorrectly Identified" divided by the "Total Number of Organisms in the Original Count", multiplied by one hundred. Samples pass if the "Percent Incorrect Identifications" is less than or equal to 5% at the family level (10% at species level). The manual provides examples (not included here) of how to calculate ‘Percent Incorrect Identifications’. Appendix E - appendices Page 26 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • For new persons, projects, or sampling runs, quality control staff should check the first five samples identified. If one of the first five samples fails the QA/QC protocol, the process begins again until five consecutive samples have passed. • Following the initial five samples, a random selection of two samples in the following ten, two samples in the following 30 and two samples in the following 50, will be checked. If any of these samples fail the QA/QC protocol, the person must again pass five consecutive samples. • Staff checking samples will have adequate experience in identification. Where possible, QA/QC of sample identifications should be conducted by more than one person to avoid bias and increased workloads. All misidentifications will be shown to the person and suitable instruction given to rectify the mis-identification. Other samples containing taxa that were misidentified are then checked for identification errors by the original identifier. Laboratory sample record sheets are to be fully completed by the person conducting the QA/QC check. Error/action codes are to be recorded on laboratory sample record sheets if appropriate. Victoria (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/Vic/) • Prior to a field trip, the number of samples to be taken in the catchment(s) visited during the trip shall be determined. Coloured marbles equal to this number, plus additional white marbles greater than or equal to 10% of the number of expected samples are to be added to a jar. Immediately after sampling and live-picking has been conducted at each site a marble shall be randomly selected from the jar for each sample. Marbles are not returned to the jar after selection. If the marble is other than white, no residue sample need be collected. If the marble is white, the sample residue remaining after live-picking shall be collected in a large, sealable container, preserved in 80% ethanol solution and returned to the lab. • EPA will retain five per cent of residue samples for internal quality control of live-picking efficacy. Macroinvertebrate samples • Each residue sample shall be randomly sub-sampled using a box subsampler and ten per cent recovered for picking and identification of macroinvertebrates under a dissecting microscope. Macroinvertebrates shall be identified to family as for the live-picked samples and where taxonomic resolution allows. Results from the residue samples will be recorded and entered onto the EPA biological database under method code 31 for sweep sample residues and method code 32 for kick sample residues. • For quality control purposes, and in particular to ensure consistency across laboratory staff, a random selection of 10% of all sorted macroinvertebrate samples shall be re-identified by a senior taxonomist or ecologist. Errors at family level should be less than 1% and errors at species level should be less than 10%. Sample processors are responsible for ensuring that their identifications are checked for quality. • For each sample processor, quality control checks shall be undertaken on 10% of samples identified to species level and independently on 10% of Appendix E - appendices Page 27 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report samples identified to family. Samples from different catchments can be grouped for QC testing, providing that all samples have been identified by the same processor at one level of taxonomic resolution. • The checker shall select in a random process the sample(s) that will be quality controlled. Appropriately qualified staff shall undertake quality control: • For all discrepancies between processor and checker identifications, an appropriate third person shall be consulted and a consensus decision made on the actual identification. • The checker will complete the standard QC report form and discuss the results with the processor. The form shall then be placed into the laboratory QC folder, and details of the QC check entered onto the front index page of the folder. Sample sorting • As a minimum, 10% of all sample residues are to be preserved for quality assurance/quality control assessment, using the same procedures (envelopes etc) as conducted under MRHI Phase 1 rounds 3 and 4. Sample selection is organised by the lead agency. Stratified random allocation of samples must be conducted with respect to operator, catchment and habitat. The person with knowledge of specific sites/operators for which samples require preservation must not be involved in sample sorting. Half of these samples (ie. 5% of the total sample number) must be sorted and an assessment made of individual operator quality assurance/quality control performance. A proportion of the remaining samples must be forwarded for external quality assurance/quality control (to be arranged). • Additional internal quality assurance/quality control is to be conducted as agreed with the Program Coordinator. Taxonomy • 5% of all processed samples, accompanied by their data sheets, must be forwarded for external quality assurance/quality control for taxonomic identification (to be arranged). Training • All staff involved in live picking must be provide with supplementary basic training in the above modified sampling techniques proper to conducting any field sampling for the assessment. Tasmania (1) (Krasnicki et a 2001) NOTE: The following information has been taken from Krasnicki et al. (2001) which documents the internal QA/QC programs that were run from the start of the MRHI program through to the end. It is unknown whether indicated QA/QC guidelines and procedures are/were applied during AusRivAS sampling. To this effect, the QA/QC procedures documented in the Tasmanian AusRivAS sampling manual, which are far less comprehensive than those described by Krasnicki et al. (2001), are listed separately under Tasmania (2). Appendix E - appendices Page 28 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report As outlined in Krasnicki et al. (2001), quality assurance/quality control procedures are designed to establish an acceptable taxonomic standard of macroinvertebrate sorting and identification. The quality control component is to determine the variation in the level of identifications, and quality assurance provides potential users with the assurance that the accuracy of results is within controlled and acceptable limits. The aim of the internal QA/QC program was to assess individual performance, detect problems at an early stage and thereby allow intervention and training to proceed before the quality of the data gathered in the program was seriously compromised. Rapid assessment sampling and site/habitat assessment • Ecologists involved in the MRHI program attended a training day that involved demonstration of the sampling protocol and practice sessions prior to the first round of sampling (Spring 1994). Any staff involved in the sampling process since then have also undergone similar training in order to standardise the MRHI sampling protocol. • New staff were trained by experienced staff in the appropriate rapid assessment protocol and environmental variable assessment techniques developed for NRHI sampling. This process involved visiting a range of sites around Hobart, where ecologists sampled riffle and edgewater habitats and performed live picks and completed all aspects of the habitat data sheets. This allowed sampling techniques to be standardised according to the River Bioassessment Manual (CEPA, 1994) and live-picking protocols and also enabled staff to compare and standardise physical descriptive techniques according to the Tasmania AUSRIVAS Sampling and Processing Manual. Upon commencement of fieldwork, the two field teams also assembled at several sites and cooperatively sampled the sites covering a range of habitats from upland streams to lowland rivers, giving the staff experience in sampling from a wide range of river types and was used to identify potential problems and sources of error associated with the sampling regime. Identification • All staff employed on the MRHI program had prior experience in macroinvertebrate identification • Initial training for ecologists was given in a two-day taxonomic workshop held by the Murray-Darling Freshwater Research Centre. Identification sessions were held by the relevant taxonomic experts to train participants in the use of taxonomic keys and to assist in identification of specimens from difficult groups. • Ecologists associated with the MRHI program have attended these workshops and have passed on knowledge gained in the use of new macroinvertebrate keys to all staff involved in identification. • DPIWE laboratory uses a reference collection of the most recent keys from these workshops in macroinvertebrate identification. • The number of ecologists involved in identification was kept to a minimum where possible to help reduce misclassification errors. All staff worked in the same laboratory and any questions were encouraged and dealt with at the time by experienced biologists on hand or at the Zoology Department, University of Tasmania. Appendix E - appendices Page 29 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Approximately 5% of samples collected from all sampling rounds were cross checked. Samples were systematically selected to cover a range of biogeographical regions, habitats and staff. • Internal process for QA/QC for macroinvertebrate identifications consisted of staff experienced in taxonomic identification of macroinvertebrates reidentifying and counting the selected samples and then cross checking the results against those provided by the original staff. A miss-identification error of <10% of the total number of animals was deemed acceptable at the family level. • Identification issues were resolved with additional training or updating of staff in taxonomic issues. Past samples were rechecked and where appropriate the database was updated. Rapid assessment live picking efficiency • For two sampling rounds, a few sites were sampled intensely by two operators to monitor live picking efficiency in the field. Each operator’s sampling efficiency was compared both within the riffles at each site and between the two samplers, with the aim of identifying any taxa that were consistently missed. As this method was time-consuming and representative of limited conditions it was not continued through the remainder of the MRHI program. • The QA/QC process involved preserving a minimum of 10% of all sample residues, both riffle and edgewater, from each sampling round. Samples were selected to cover a broad range of biogeographical regions, habitats and staff. Half (5%) were processed internally to assess operator sorting efficiency and the other halt were sent to be assessed externally. Analysis was carried out as per previous external audits conducted by ERISS (Bray Curtis dissimilarity values and Live Sort/WSE ratios). Site and Habitat Assessment • This part of the sampling procedure was performed by two ecologists whose job was to solely describe and record environmental variables. To minimise the effects of operator error an attempt was made to keep staff who performed the habitat assessments constant and this was maintained for the Autumn 1995 to Spring 1997 sampling rounds. Subsequent rounds usually then had at least one experienced staff member who was able to train any additional staff required to perform site and habitat assessment. • To assess consistency of data recorded, ecologists from two of the rounds were required to independently assess site and habitat parameters at the same site, from a number of NRHI locations. Variables assessed were those that required some element of subjective judgement (eg. substrate composition). Ecologists new to the program were trained in all aspects of habitat assessment incorporating those areas that were highlighted as potential for operator error. Data Entry • All biological and environmental data were checked for entry errors prior to performing the final classifications. • A person independent of the data entry examined the data and cross checked it against the data sheets. Any missing or incorrect data was corrected. Appendix E - appendices Page 30 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Additional checks were performed by plotting biological and environmental variables are frequency histograms. Outliers were examined to establish whether they were in the correct range and the independent data checker also had the task of judging whether the data, even though it fell within the reasonable ranges, was still appropriate or usual for that type of site. This meant that the checker had a working knowledge of the data set. Tasmania (2) (AusRivAS Manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/Tas/) After picking, 10% of the residues are randomly selected and preserved in formalin for QAQC analysis of operator sorting efficiency. Sorting The QA/QC program aims to assess the effectiveness of individual operator sorting procedures using as its basis, comparison of the composition of live-picked samples with associated residues. The MRHI Bioassessment manual states that the aim of the live pick procedure is to ensure that the broadest range of biota are collected at a site. This implies that the taxa list derived from a live pick will encompass more taxa than would be expected if a random sample of animals of equivalent number to the live sort total were drawn from the sample (ie. ‘whole sample estimate’). • Approximately 10% of all riffle and edgewater residues are to be preserved from each sampling round. Half of these (5%) are to be processed so that operator sorting efficiency can be assessed • Analysis is carried out as per previous external audits conducted by ERISS Identification • Approximately 5% of the samples collected each round are cross-checked by persons with adequate identification experience. • Samples are selected to cover a broad range of biogeographical regions, habitats and staff. • A miss-identification error of < 10 % of the total number of animals is deemed acceptable at the Family level. this is the error rate used by the Murray Darling Freshwater Research Centre who conducted external quality control checks of all State agencies • In all cases, identification problems are to be resolved with additional training of staff. Past samples containing taxa that were found to be misidentified are to be rechecked and, where appropriate, the database updated. South Australia (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/SA/) Field sampling • The South Australian program has involved a small group of biologists from the same laboratory collecting all samples from 1994 to 2000. This has Appendix E - appendices Page 31 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report ensured that an experienced operator has been involved with sampling each site, and over time all members in the group have gained considerable experience sampling a wide variety of river types throughout the State. This has ensured a consistent approach to identifying habitats and field sampling effort during the course of the program. • It is recommended that any future use of this protocol should involve the participation of at least one of the team members involved with this work at the Australian Water Quality Centre or EPA to minimise the chance of introducing significant sampling errors. Laboratory • The sub-sampler is subject to an internal check of an additional 5% of samples to ensure random sub-samples are being produced with the laboratory sorting method used in S.A. • In addition, ERISS carried out external checks of residues from the 1995 surveys to test the performance of the sub-samplers used by each state and territory (where laboratory sorting methods are used). That work confirmed the sub-sampler operated effectively to randomly sort the sample and allow the retrieval of a representative 10% sub-sample. • A 5% randomised selection of residues from each survey has been kept in storage for any possible future QA of the sorting protocol used in S.A. • As part of a national QA/QC program involving the identification of macroinvertebrates, the Murray-Darling Freshwater Research Centre independently checked samples that had been sorted and identified by the team in S.A. Samples were assessed for the 1994, 1995 and 1997 surveys. The results from these showed the high performance of the approach used in S.A. and indicated that no further work was needed to improve the identification of specimens from this State. • All new staff are trained in the use of the sub-sampler and identification keys used in S.A. The experienced team members have also assisted new staff to identify organisms that they are not familiar with and check difficult taxa. • The addition of new staff during the program led to the development of a more rigorous internal training protocol in 1998. This included: à à à Random checks of sorting trays of new members to ensure all specimens were being collected and more importantly that novel taxa were not being overlooked. All staff involved with the project process and identify a contrived sorted sample to provide a check on counting and identification skills. Random checks of identifications carried out by all operators. Western Australia (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/WA/) • As a minimum, 10% of all sample residues are to be preserved for quality assurance/quality control assessment • Stratified random allocation of samples must be conducted with respect to operator, catchment and habitat Appendix E - appendices Page 32 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • The person with knowledge of specific sites/operators for which samples require preservation must not be involved in sample sorting. • Half of these samples (ie. 5% of the total sample number) must be sorted and an assessment made of individual operator quality assurance/quality control performance • Additional internal quality assurance/quality control should be performed Northern Territory (AusRivAS manual: http://ausrivas.canberra.edu.au/Bioassessment/ Macroinvertebrates/Man/Sampling/NT/) • Quality control / quality assurance procedures are designed to establish an acceptable standard of macroinvertebrate sorting and taxonomic identifications. The quality control component minimises the variation in sorting and identification efficiency. Quality assurance provides potential users with the assurance that the accuracy of results is within controlled and acceptable limits. Sorting Efficiency • All new staff are to be trained in the NT laboratory procedures including subsampling, sorting and sample storage. • Once sorting has commenced, the residues of the first five samples are checked by quality control staff. • An assessment is made following the first five samples whether sorting efficiency is acceptable. • If not satisfactory the checking is continued for every sample until the problem is rectified. • If the sorting for the first five samples is acceptable a random selection of two samples in the following ten, two samples in the following 30 and finally 1 sample in every 20 will be checked by quality control staff. • The protocol is repeated for the commencement of new projects and sampling runs as well as new staff. • A sorting efficiency of > 90% is deemed acceptable. Sorted samples are resorted and missed animals are identified, enumerated and compared to the animals collected from the original sort. If the number of animals of a particular Family counted in the re-sort is ?10 % of the total count (original plus re-sort count for that Family), the person is given suitable instruction to ensure that particular Family is adequately collected in future samples. • It is important when sorting to ensure all different taxa in a sample are collected. If a particular Family contributes significantly to the count (> 10% of the total count) an error in the count of ?10% is less important than in a Family with a lower count. Where errors occur in the count of Families which contribute significantly to the total count, sorting staff are cautioned and made aware of the these groups to reduce the chance of significant errors in future samples. Taxa not collected in the original count but collected in the re-sort are treated as a significant error with the appropriate instruction to correct the problem. Appendix E - appendices Page 33 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Identification and Enumeration Efficiency • All new staff are to be trained in the NT laboratory procedures including subsampling, sorting, identification, enumeration, sample storage and archiving. • Once sample identification has commenced, the identification results of the first five samples are checked by quality control staff. • An assessment is made following the first five samples whether identification and enumeration accuracy is satisfactory. • If not satisfactory appropriate instruction is given to the person as described in the boxed section below. • If identification and enumeration for the first five samples is acceptable a random selection of two samples in the following ten, two samples in the following 30 and finally 1 sample in every 20 will be checked by quality control staff. • The protocol is repeated for the commencement of new projects and sampling runs as well as new staff. • A misidentification error of < 10 % of the total number of animals is deemed acceptable. This is the error rate used by the Murray Darling Freshwater Research Centre who conducted external quality control checks of all State agencies participating in the AusRivAS program. If the error is > 10 %, misidentifications are corrected under the guidance of quality control staff. All misidentifications will be shown to the person and suitable instruction given to rectify the misidentification. Other samples containing taxa that were misidentified are then checked by the original identifier for misidentification errors. • When an identification problem is encountered, a decision tree for identifications should be followed as is figured in Hawking and O’Connor (1997). • Very small, damaged, immature animals or pupae that cannot be identified with confidence should be noted as such (eg. Trichoptera juvenile). These animals are counted and placed in separate vials for each category. The counts for unidentified animals are not included in the 200 organism subsample. • Damaged animals should be identified if possible, with both head and tails counted and the highest number recorded and placed in the appropriate vials. If a specimen cannot be identified it should be noted as such (eg. Ephemeroptera damaged) and placed in the appropriate vial. The same procedures apply to the identification of Oligochaeta. • When identifying the samples, the taxa are separated into Orders and placed in separate vials to eliminate any high level discrepancies. This is also required for future curatorial preservation and storage. Appendix E - appendices Page 34 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Appendix E:7 Live Picking Methods These descriptions of live picking methods are derived directly from each state / territory methods manual. Two of the State protocols are described in flowchart form below to illustrate the methodology (diagrams from WATER ECOscience, 2003). 1 Queensland Protocol Victorian Protocol Appendix E - appendices Page 35 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Queensland • It is recommended that you initially separate small organic and substrate material from large using a 1 cm panning sieve. Sort through these fractions a small amount at a time, retaining the residue for QA/QC requirements. Pick progressively through the sample (using forceps and/or pipette), replacing picked material with remaining parts of the sample as picking progresses, until a total of 200 animals have been collected or for 60 minutes (whichever is completed first) attempting to pick as many different groups (families) as possible. • Collect a maximum of 10 of any one type (family and in some cases order) or animal. At least 20 midge larvae (Chironomidae) should be collected to ensure adequate representation of the subfamilies. • For the first 15 minutes, collect from the predominant groups, remembering only 10 animals per group • For the next 30 minutes scan for cryptic and/or rare groups. These will tend to be the following groups: → Corbiculidae (juveniles → Chironomidae (larvae and pupae) → Empididae → Hydroptilidae (larvae) → Ceratopogonidae (larvae) → Oliogchaeta (including broken bits) → Elmidae (larvae) → Hydrophilidae (larvae and adults) → Simulidae (larvae) • For the final 15 minutes return to the more common animals • If it is raining or cold, or conditions of poor light exist due to cloud cover or approaching twilight, the sample must be taken back to the vehicle/motel/camp etc. for sorting undercover and with improved light conditions. Laboratory sorting • Ensure adequate ventilation in the workplace. Tip the contents of the jar into a large petri dish; or pour the contents through a 250 um sieve over a sink, wash the organisms and flush the sieve contents into a large petri dish with water from a squeeze bottle • Place the petri dish under a stereomicroscope. Take a vial of suitable size to take the collection of specimens in the petri dish with label inserted. The label should have the following information: collection number, location code, site name, collection date, habitat, sample identifier, and the identification date. • A dedicated tally sheet should be developed for recording the identities and numbers of all taxa in a sample (see appendix). The sheet should allow listing of the taxonomic key used for identification for each family, the person making the identification, the site, date and sample code. Appendix E - appendices Page 36 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • Organisms are identified to family level except for Porifera, Nematoda, Nemertea, Oligochaetes, Acarina and microcrustacea for which family level identification is optional. Chironomids should be identified to subfamily level. • Select specimens and follow the appropriate taxonomic keys to family level. Follow the keys to family level. If uncertain about the identity obtain a second opinion from a colleague/local specialist. If a new family is suspected or other significant problems arise in taxonomic identification, contact a national specialist. • Identify each specimen, place in the vial and mark the tally sheet. When all specimens have been counted record the total tally for each taxa. Place the vial, filled with preservative, in an evaporation proof box in a suitable storage location. Victoria • The sample is washed in the net and then spread out onto a large white tray where it is sorted for a standard time between 30 and 60 minutes. Animals are collected with the aid of forceps, pipettes and spoons. Many animals can be quickly collected from the corners of the tray, particularly when using pipettes. Once or twice during the sorting it is advisable to strand animals by tilting the tray to one side, thus exposing a third or half of the bottom. Rapidly moving animals can be collected in this way, as well as molluscs, which adhere to the bottom of the tray. • If large amounts of leaves, wood or aquatic vegetation are collected, these should be rinsed and removed before sorting. If the water is cloudy due to clays or fine sediment in suspension, put the sample back into the net and rinse it in the stream again. • If large amounts of sand or coarse organic material are collected, put only a proportion of the whole sample into the tray at one time, bearing in mind that you need to completely sort the whole sample within the allotted time. • Good practice requires working in relatively high light levels. If ambient light is low then artificial lighting will be required. Rain drops also adversely affect sorting ability and umbrellas or tarps should be used. • The main objective of sorting is to collect as many different taxa as possible. Care must be taken not to take too much time picking out large numbers of very abundant species, as this will result in the less common species being under represented or not collected. Only about 30 of each taxa need to be picked out, they can then be ignored and the remaining sorting effort applied to collecting other species. Considerable effort needs to be directed towards searching for small or cryptic species. • The number of animals collected in 30 minutes is typically about 200, grossly impacted sites are likely to have fewer than 50. The live sorting should aim to collect about 200 animals in the allotted time. If after 30 minutes fewer than 100 animals have been collected, sorting should continue. If no new taxa are found in the next ten minutes, cease sorting. If new taxa are found continue up to a maximum of 60 minutes. Appendix E - appendices Page 37 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report Laboratory sorting • The sample is preserved in 80% ethanol and returned to the laboratory for identification. • All identifications should be carried out with a stereomicroscope using appropriate keys and up-to date texts. For a listing of invertebrate keys, see Hawking (1994). Animals should be identified to family level with the exception of Oligochaeta, Hydracarina, Chironomidae (subfamily), Hirudinea, Platyhelminthes. Western Australia • Empty the contents of the net into a bucket of water and swirl vigorously to separate the mineral substrate from the organic component of the sample. Pour the water fraction onto a stack of four sieves. The four sieves should have mesh apertures of 16mm, 2mm, 500 um, 250 um, with the top sieve having the coarsest mesh size progressing down to the 250 um mesh sieve at the bottom of the stack. Continue to elutriate the sample in this manner until confident that all animals have separated from the mineral component of the sample. After elutriating the sample, agitate the stack of sieves in the water column to separate the contents into the four size fractions. Empty each of the four size fractions into separate sorting trays. Separating the samples into the various sizes fractions facilitates the picking process by refining the search image of the collector. • The sample must be picked by two collectors for a total of 60 mins ie. 30 mins by each collector. Allocate picking effort to each tray proportional to the amount of material it contains. Typically the larger fractions (16mm and 2mm) are picked for longer (eg. 9 minutes by each collector) with less time being allocated to the smaller fractions (500 um and 250 um – 6 minutes each). • Remove animals from the trays using forceps and /or a pipette and store in a vial containing 70 % alcohol. • The aim of the sampling is to maximise the diversity of the animals collected. Start by collecting common, abundant taxa for the first five minutes. After that the major picking effort should be directed at finding the less common, inconspicuous taxa. Avoid over-picking large or colourful taxa. Aim for a total of 200 animals (use a hand held counter) with maximum diversity. There is no need for large numbers of any single taxon but a minimum of 30 chironomids should be picked from every sample to ensure that all the subfamilies are represented in the vial. • Particular care should be taken to search for the groups that can be commonly missed when live sorting (ie. cryptic taxa) : elmid larvae, Oligochaeta, Empididae, Hydroptilidae, small molluscs, Ceratopogonidae • If it is a really poor sample with very few animals in total, then stop at 60 mins. Make it clear on the field sheet that it is a poor site and why that was so. Laboratory Procedures • Tip the contents of the vial into a large petri dish. Place the petri dish under a stereomicroscope which is correctly adjusted for your vision and work posture. Use a vial of suitable volume to store the specimens in the petri dish. Label the vial with the following information: site code, site name, Appendix E - appendices Page 38 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report collection date and habitat. Use a pencil or waterproof ink to fill in details. Store the specimens in 70% ethanol. • A dedicated tally sheet should be developed for recording the identities and numbers of taxa in a sample. (see appendix) The sheet should allow listing of the key used for identification for each family the person making the identification the site and date and sample code. • Organisms should be identified to familiy level with the exception of Oligochates, Hydracarinids, • Ignore all microcrustacea (ostracods, copepods, cladocerans) but include conchostracans and anostracans. Chironomids should be identified to subfamily level • Follow the keys to family level. If uncertain about the identity obtain a second opinion from a colleague/local specialist. If a new family is suspected or other significant problems arise in taxonomic identification, contact a national specialist. • When each specimen has been confidently identified, mark one stroke in the tally column of the data sheet, place the specimen in the vial and examine the next specimen. When all specimens have been counted, add the tally for each taxon and write the total in the appropriate column on the data sheet. Place the vial, filled with preservative, in an evaporation proof box in a suitable storage location. Northern Territory • Whole samples are preserved in 90% ethanol and taken back to the laboratory for processing. Laboratory procedure • Set up a 250 um mesh sieve in a large container. Place the sample in the sieve and allow the ethanol to drain into the container. Remove and save the ethanol. Rinse the sample pot containers and lids with water into the sieve. Refill washed containers with recycled ethanol. Rinse the sample and wash out any fine sediment. • Place the sample into a subsampling device. In the NT a waterproof ‘Pelican’ case is set up with a perspex rack containing small plastic vials. A fitted wire mesh is used to hold the vials and perspex rack in place. This type of box subsampler is based on a subsampler designed by Marchant (1989) • Wash the sample into the subsampler box and fill with water until the case is full. Lift the case up and invert so that the sample always falls away from the vials. Agitate the case vigorously while inverted. While the sample material is still being agitated flip the case upright onto the bench top. This should ensure the sample is evenly distributed throughout the vials in the box. • A 200 organism subsample is required. Randomly select vials from the case and place into a rack. Sort samples under a stereo microscope and remove and count all macroinvertebrates. Record the number of vials required to obtain the 200 organisms. Extracted vials must be completely sorted even if the 200 organism count is reached, to enable estimates of total numbers. Appendix E - appendices Page 39 WATER ECOscience: QA/QC Project – Year 2 Milestone and Final Report. Appendix E: Macroinvertebrate Sample Processing Error Report • All macroinvertebrates are to be identified to family level except for the following, using the keys recommended by Hawkings (1999). Adults and larvae are combined for the purpose of data entry and analysis. → Oligochatea (Class) → Acarina (Order) → Chironomidae (sub-family) Appendix E - appendices Page 40
© Copyright 2026