Document 377376
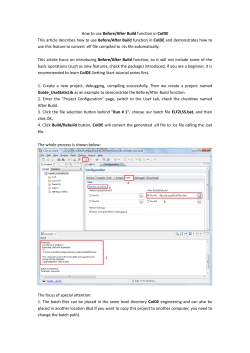
Predic'ng Signal Pep'des using Deep Neural Networks Cecilie Anker, Casper Sønderby and Søren Sønderby Introduc'on Central Dogma DNA RNA Transcrip7on Training examples 600000 479175 400000 200000 0 37466 1640 23917 SP CS TM Signal pep'de (degraded) Mature protein (secretory pathway) Cleavage Site TTGNGLFINESAKLVDTFLEDVKNLHHSKAFSINFRDAEEAK! SSSSSSSSSSSSSSSC....................TTTT..! Slide window across sequence Neural network Input window ENCODINGS TABLE 1! Other Figure 2: Composition of dataset. Signal peptide (SP), Cleavage site (CS), Trans membrane (TM). ! 000001000010000000000001000000010000000001! Figure 3: Schematic overview of the encoding method used. Signal peptide (red), mature protein (blue). Data is encoded using a sliding window where each amino acid is encoded using one of the encodings schemes from table 1.! TABLE 1: Different feature encodings that were evaluated. Asterisk indicate feature encodings that were used in the DNN network ensembles.! DNN Probabili7es HMM-‐DNN hybrid Model HMM-‐DNN DNN model output Figure 4: Schematic view of nested cross-validation DNN ensembles and DNN-HMM hybrid model. In the HMM-DNN hybrid model the HMM emission probabilities are equal the DNN output.! Results The best performing ensemble of networks used a combina7on of both sparse and NLF encoded data. None of the other encoding schemes were found to increase performance. Inclusion of networks using different window sizes and network architectures were also found to increase the ensemble performance. One of the four ensembles are shown in table 2. The Performance of the DNN and HMM-‐ DNN hybrid are shown in table 3 as MCC scores. For comparison the scores of the neural networks used in signalP 4.0, as reported in Petersen et. al [1], are shown although the exact composi7on of the test set is not iden7cal why no direct comparison can be made. The DNN performs best on all performance measures, except SP MCC where the HMM-‐DNN hybrid models has best performance. Architecture Dataset Window Op'miza'on 882-‐600-‐600-‐600-‐4 Sparse 41 66% cs sens, 33% cs prec 672-‐600-‐600-‐60-‐4 Sparse 31 50% cs sens, 50% cs prec 652-‐300-‐300-‐30-‐4 NLF 31 66% cs sens, 33% cs prec 652-‐1200-‐1200-‐1200-‐4 NLF 31 SP MCC 672-‐1200-‐1200-‐1200-‐4 Sparse 41 66% cs sens, 33% cs prec TABLE 2: One of the four DNN ensembles selected in nested cross-validation training ! Discussion The performance of the DNN network ensemble were found to be higher than the performance of the neural networks used in SignalP 4.0. SignalP 4.0 implements both a heuris7c scoring func7on and network selector to improve the final performance. Using this method SignalP 4.0 performs be\er than DNN’s presented here. These methods were not studied, but could poten7ally increase the performance of the DNN’s presented here. The HMM-‐ DNN hybrid model precisely predicted SP on the expense of CS predic7on performance. We speculate that higher order HMM’s might increase the performance. However the fixed simple sequence of states (signal pep7de -‐> cleavage site -‐> other) might hamper the performance gains of the DNN-‐ HMM hybrid model. Due to the high dimension of the meta parameter space it could not be densely sampled and hence human interven7on was needed. This might not be an effec7ve search strategy and the algorithm may be further improved by exploring the parameter space automa7cally using e.g. Gaussian processes [11]. Since only limited labeled data are available (especially sequences with signal pep7des) pre-‐training using the vast amount of unlabeled sequence data might increase the performance of the algorithm. This is however computa7onally very expensive. Conclusion DNN and HMM-‐DNN models for predic7ng signal pep7de CS were implemented on GPUs using MATLAB. The DNN performed be\er than the neural networks used in SignalP 4.0, although heuris7cs improvements gives signalP 4.0 be\er performance. Further studies are needed to implement these heuris7cs using the DNN and HMM-‐DNN models. References Window Nr. Widths Features 31*, 41* 672, 882 Sparse encoding [2]* Non-‐Linear Fisher transform of 31*,41* 652, 842 physiochemical proper7es(NLF) [3]* Principal component Analysis of 41 883 physiochemical proper7es [3] Blossum90 [4] 41 924 Codon encoding [5] 31, 41 760 DNN 5-Ensemble! Feature encoding: The amino acids were encoded using a sliding window with several different encoding schemes as seen in table 1. Amino acid sequence composi7on, rela7ve sequence posi7on and sparse encoding of the -‐1 and -‐3 posi7ons were added to all encodings schemes. Only the 70 first amino acids in each pep7de were considered. The targets were encoded as mutually exclusive classes. Network Architecture: 3-‐hidden-‐layer networks with sigmoid ac7va7on func7ons and soimax output were used. Network Training: The networks were trained using back propaga7on and cross-‐entropy error. The networks were regularized using dropout (input 0 or 20%, hidden 50%) and restraining the L2-‐norm of the incoming weights to the individual neurons [6]. The parameter space (feature encoding, window size, learning rate, learning rate decay, dropout frac7on, L2-‐norm and hidden layer sizes) were sampled explora7vely guided by human interven7on. The parameter seYngs of the 15 networks best predic7ng the CS Ma\hew correla7on coefficient (MCC) were used for 5-‐ fold nested cross-‐valida7on. During training each network was saved with regard to 8 different performance measures. The training of a single network took between 24 and 48 hours. Network Ensemble: An ensemble of 5 networks were used. 5-‐ensembles were chosen by evalua7ng all permuta7ons of the 38 best performing networks. The arithme7c mean predic7ons of ensembles were used for evalua7ng the test performance. HMM-‐DNN hybrid: A HMM-‐DNN model was trained, were the emission probabili7es were calculated by the DNN network ensemble [7]. Performance: The DNN was evaluated as a separate model and secondly a HMM-‐DNN hybrid was evaluated. Descrip'on Transla7on Figure 1: The central dogma states that DNA (genes) is transcribed into RNA which is translated into proteins.! Method Dataset: Protein sequences with experimental evidence for signal pep7des were used as posi7ve data (n=1640). Nuclear (n=5133) and trans-‐membrane (n=687) proteins were used as nega7ve data. See Petersen et. al for further details [1]. Protein Gene7c informa7on is stored in DNA. DNA is transcribed into RNA, which is translated into an amino acid strings, which then fold into mature proteins. See figure 1. The unfolded amino acid sequence of proteins targeted for the cell membrane and cell organelles contains a N-‐terminal signal pep7de (SP) which control the entry to the secretory pathway. The SP is cleaved from the immature protein in the endoplasmic re7culum at a specific posi7on denoted the cleavage site (CS). Hence the SP is not a part of the mature func7onal proteins making precise predic7on of the SP important for both in vitro protein synthesis and func7onal annota7on of proteins. See figure 2, top. Current state of the art methods for predic7on of CS use conven7onal shallow neural networks. This study applies deep neural networks (DNN) to the predic7on task and test recently developed regulariza7on methods preven7ng overfiYng. Furthermore a Hidden Markov Model – DNN (HMM-‐DNN) hybrid model is developed and the performance of this model is evaluated. (method con7nued) For the HMM-‐DNN the output from the trained DNN ensembles were used as emission probabili7es in the HMM. Implementa'on: Based on the the MATLAB DeepLearn-‐ Toolbox [8] a memory efficient DNN GPU implementa7on [9] and a HMM-‐DNN hybrid model was implemented [10]. Method \ Score SP CS CS CS MCC sens. Pres MCC TM MCC SignalP 4.0 NN 0.881 NA DNN 0.888 0.691 0.712 0.701 0.813 NA 0.648 NA HMM-‐DNN viterbi 0.921 0.669 0.627 0.646 0.793 TABLE 3: Test performance of DNN and HMM-DNN compared to networks used in SignalP 4.0. ! [1] Petersen, TN., et. al. (2011) SignalP 4.0: discrimina7ng signal pep7des from transmembrane regions. Nature methods 10(8) ,785-‐786. [2] Hinton, GE., et al. (2012) Improving neural networks by preven7ng co-‐adapta7on of feature detectors. arXiv preprint arXiv:1207.0580. [3] Qian, N., et. al. (1988). Predic7ng the secondary structure of globular proteins using neural network models. Journal of molecular biology, 202(4), 865-‐884. [4] Nanni, L et. al. (2011). A new encoding technique for pep7de classifica7on. Expert Systems with Applica7ons, 38(4), 3185-‐3191. [5] Wu, C. H et. al. (Eds.). (2000). Neural networks and genome informa7cs (Vol. 1). Elsevier Science. [6] Zamani, M., et al. (2011). Amino acid encoding schemes for machine learning methods. In Bioinforma7cs and Biomedicine Workshops (BIBMW), 2011 IEEE Interna7onal Conference on (pp. 327-‐333). IEEE. [7] Bourlard, H et. al, (1994), Connec7onist speech recogni7on: a hybrid approach. Springer. [8] Palm, RB. (2012), Predic7on as a candidate for learning deep hierarchical models of data, master thesis. [9] h \ p s : / / g i t h u b . c o m / s k a a e / D e e p L e a r n T o o l b o x / b a t c h v a l [ 1 0 ] h\ps://github.com/skaae/hmm-‐hybrid. [11] Snoek, J. et. al. (2012). Prac7cal Bayesian op7miza7on of machine learning algorithms. arXiv preprint arXiv:1206.2944.
© Copyright 2026