transplantation immunoglobulin repertoire during
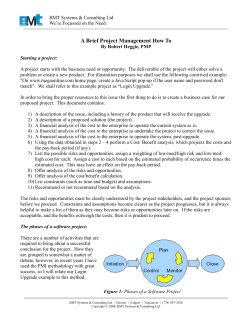
From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 1996 87: 2795-2804 Evidence for oligoclonal diversification of the VH6-containing immunoglobulin repertoire during reconstitution after bone marrow transplantation I Nasman and I Lundkvist Updated information and services can be found at: http://www.bloodjournal.org/content/87/7/2795.full.html Articles on similar topics can be found in the following Blood collections Information about reproducing this article in parts or in its entirety may be found online at: http://www.bloodjournal.org/site/misc/rights.xhtml#repub_requests Information about ordering reprints may be found online at: http://www.bloodjournal.org/site/misc/rights.xhtml#reprints Information about subscriptions and ASH membership may be found online at: http://www.bloodjournal.org/site/subscriptions/index.xhtml Blood (print ISSN 0006-4971, online ISSN 1528-0020), is published weekly by the American Society of Hematology, 2021 L St, NW, Suite 900, Washington DC 20036. Copyright 2011 by The American Society of Hematology; all rights reserved. From www.bloodjournal.org by guest on December 3, 2014. For personal use only. Evidence for Oligoclonal Diversification of the VH6- Containing Immunoglobulin Repertoire During Reconstitution After Bone Marrow Transplantation By Ingrid Ngsrnan and lnger Lundkvist Patients who have undergone bone marrow transplantation (BMT) remain immunodeficient for months to years posttransplantation. To evaluate the basic molecular events underlying reconstitution of the humoral immune response, we have performed a detailed nucleotide sequence analysis of VH6 containing rearrangements in circulating B cells from two BM donorlrecipient pairs. Our results show that the third complementarity determining region (CDR3) diversity is much lower early after transplantation, compared with that of the donors, and that the clonal variability remains low for 3 months. Repertoire diversificationfollows an oligoclonal pattern where B lymphopoiesis appears to occur in waves up to 6 months posttransplantation. The repertoire among donated marrow cells is not reflected in peripheral blood lymphocytes fromthe transplanted patients. There is differential D geneutilization among both donor andpatient samples, whereas JH gene usage is biased toward Jn4, 5, and 6. One month after transplantation the vast majority of the sequenced clones are functional and contain a high frequency of replacement mutations in the CDRs of the v ~ 6 gene. We conclude that lg gene expression is very restricted early after B M 1and that development of the B-cell repertoire appears to follow a wavelike pattern. 0 1996 by The American Society of Hematology. I frequently expressed family is vH3. Interestingly, vH3 gene family usage in the fetal repertoire is not random but three genes, 2 0 ~ 1 ,30p1, and 5 6 ~ 1 ,are markedly overrepre~ented.’~.’~ The vH3 family also dominates the repertoire among peripheral B lymphocytes from healthy a d ~ l t s . ~ ~ ~ ’ ~ Among naturally activated lymphocytes, however, the VH6 gene is overrepresented while expression of the vH1 and vH3 gene families is decreased (Davidkova G, Pettersson S, Holmberg D, Lundkvist I: Selective usage of VH-genes in adult human B lymphocyte repertoires. Submitted for publication). Fumoux et al’’ have analyzed VH gene family usage in BMT patients and found that vH3 utilization is decreased twofold to threefold, compared with normal adults, and is compensated for by transient overexpression of vH4, vH5, and VH6.This bias in VHgene family usage is most significant 60 days after transplantation, although the pattern is obvious also at 30 and 90 days posttransplantation. It has also been reported that the vH3 genes that characterize the fetal repertoire, 56pl and 2 0 ~ 1as , well as the v H 6gene are markedly overutilized 2 to 5 months posttransplantation.2n At 21 months after BMT the percentage of B cells that use these genes are still twofold to sixfold higher than in healthy adults.” The importance of the VH6 gene in fetal and neonatal VH gene repertoires, together with the similarities between ontogenic development and the posttransplantation humoral T IS WELL KNOWN THAT bone marrow (BM) transplant recipients develop a cellular and humoral immune deficiency that can last for months to years after the transplantation. The time required for recovery is prolonged if the patient also develops chronic graft-versus-host disease (GVHD).’ It has been suggested that reconstitution of the humoral immune system follows ontogenic de~elopment.’.~ The first circulating IgM+ B lymphocytes are detected in the periphery 2 to 4 months posttransplantation’ and the phenotype of these B cells resembles cord blood B cell^.^,^ There are few IgG and IgA expressing B cells present after BM transplantation (BMT).3 The B-cell deficiency in BMT patient^^.^ has been suggested to be caused by defective B cells, decreased TH cell function, or a suppressive effect caused by T cells or natural killer (NK) cells.’ The serum levels of Igs are normalized 6 months (IgM and IgG) to 1 year (IgA) post BMT.’ Although total IgG levels are normal, the subclass pattern is biased against y2 and 74,compensated by an increased amount of y 1. The pattern of reconstitution of cellular subsets and serum Igs evoked the question whether the posttransplantation immune deficiency could be explained by restriction in utilization of Ig genes similar to that seen in ontogeny. A number of studies have addressed the question of expression of genes coding for the variable region of the Ig heavy chain (V,) at various time points in ontogeny and in central and peripheral lymphoid tissue. The variable and constant region genes for the human Ig heavy chain are located on chromosome 14q. The diverse repertoire of Ig specificities is obtained through a multi-step process where the initial event is recombination of one of at least 30 diversity (D) genes’ to one of six joining (JH) genes9 This D-JH complex is then recombined to one of roughly 100 VH In addition to combinatorial diversity,15junctional diversity (addition of N and P nucleotide~),“.’~ differential association with K or A light chains, and somatic mutation’’ contribute to the variability of the Ig repertoire. The human VH genes are divided into seven different families based on nucleotide sequence hom01ogy.l~ The largest family is vH3, which contains 28 gene segments with open reading frame and the smallest is V,6, with one functional gene.20 Studies on VH gene usage in the fetal repertoire show an overrepresentation of the VH6family,21.22 although the most Blood, Vol 87, No 7 (April I ) , 1996: pp 2795-2804 From the Department of Immunology, Microbiology, Pathology and Infectious Diseases, the Division of Clinical Immunology, Karolinska Institutet at Huddinge University Hospital, Huddinge, Sweden. Submitted June 28, 1995; accepted July 27, i995. Supported by grants from the Children Cancer Foundation of Sweden, Jeansson’s foundations, King Gustaf the Vth 80-Year Foundation, the Swedish Cancer Society, and the Baxter Novum Research Laboratory. Address reprint requests to Ingrid Nasman, BSc, Division of Clinical Immunology, F79, Huddinge University Hospital, S-I41 86 Huddinge, Sweden. The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact. 0 19% by The American Society of Hematology. 0006-497//96/8707-0006$3.00/0 2795 From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 2796 NASMAN AND LUNDKVIST immunesystem, haveledus to study the developmental program of Ig gene rearrangementpost BMT. To gain better insight into the molecular mechanismsunderlying repertoire expressionposttransplantation, we have collectedmaterial before and at several timepoints after BMT and performed a detailed nucleotide sequence analysis of VH6-containing rearrangements. For this purpose we haveused the polymerase chain reaction (PCR) technique with VH6-specific and JH consensus primers. The first post BMT sample is taken amount of B at 4 weeks,ie, before there isasignificant lymphocytes in the periphery. We have also analyzed samples from the BM donor to be able to evaluate the importance of the repertoire of the donated cells versus that of the microenvironment on reconstitution of the immune system. MATERIALS ANDMETHODS Patients and donors. Peripheral blood (PBL) samples were derived from two BM recipients. The first (patient l ) was a 50-yearoldwomanwiththediagnosisacutelymphoblasticleukemiaof pre-B type (pre-B ALL, LI). TheBM donor (donor 1) was an HLAidentical sibling. Patient 1 was in partial remission with 7% to 8% lymphoblasts in theBM 1 week beforeBMT.Theprotocolfor conditioning of patient I was cyclophosphamide (60 mg/kg/d) for 2 consecutive days (days S and 4 before BMT) followed by total body irradiation with a dose of 10 Gy. The lungs were shielded and I wasadministered receivedadose of 9Gy.AfterBMTpatient methotrexate and cyclosporin as prophylaxis against GVHD. 3 months. Details reCyclosporin treatment was administered for garding treatment have previously been published.2" Six days after BMT patient I developed a mild (grade I) acute GVHD manifested as a localized skin rash. Five months after BMT patient 1 developed chronic GVHD that engaged the skin and mouth cavity. Before BMT patient I was negative for cytomegalovirus (CMV), but 2months after BMT CMV was detected in blood samples by PCR, the infection was asymptomatic. Nine months after BMT patient I is healthy with no relapse. The second recipient (patient 2) was a 44-year-old man with the diagnosis lymphoma (K1 +). The BM donor (donor 2) was an HLA-identical sibling. Patient 2 was in complete remission with no detectable tumor cells in the BM I week before BMT. The protocol for conditioningof patient 2 was as described above. Patient 2 was treated with methotrexate and cyclosporin. No sign of GVHD was seen and the cyclosporin treatment was stopped 4 months after BMT. Patient 2 had IgG antibodies against CMV before BMT and theseantibodieswere still present 6 months post BMT. No IgM anti-CMV antibodies were detected in serum. There were no signs of CMVdisease.Patient2 is healthy with norelapse I 1/2 years after BMT. Serum Ig levels and white blood W// count. Values in Table I were obtained from standardized routine analysisof patient samples. Cell preparation. Mononuclearcellswereobtainedfrom both BM andPBL by gradientseparationonlymphoprep(NycomedPharma AS, Oslo, Norway). The cells were washed twice in RPM1 (GIBCO BRL, Life Technologies Ltd, Paisley,UK) without supplements and the cell pellets from PBL samples were frozen in -70°C until genomicDNApreparationswereperformed.BMcellswere used for enrichment of pre-B and mature B lymphocytes. Monoclonal antibodies (MoAhs). Fluorescein (F1TC)-conjugatedanti-Calla(CD10).cloneW8E7ofmouse Ig%/K isotype (Becton Dickinson, Mountain View, CA), was used to enrich CDIO+, CD 19' B-cell precursors. R-phycoerythrin (RPE) conjugated antiCD19, clone HD37 of mouseIgG,/K isotype (DAKO A/S, Glostrup, Denmark), was used for enrichment of mature CDlO-, CD19' B lymphocytes. Enrichment of pre-B und mature B 1ymphocyle.s from the BM sample. Aschematicrepresentation of the purification procedure is presented in Fig l . The mononuclear cell fraction was incubated with FITC-conjugated antiCD10 MoAb (10 pL/I0" cells in total 50 p L phosphate-buffered saline [PBSIbovine serum albumin [BSAl) 15 to 30 minutes on ice and washed twice in cold PBS supplemented with 0.5% BSA, 5 mmol/L EDTA, and 0.01% NaN3 (PBSIBSA). The cells were resuspended in cold PBSlBSA (80 pL/107 cells) and a secondary labeling step was performed with 20 &/IO7 cells of anti-isotypic MACS magnetic microbeads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Cells and beads were incubated together for 15 minutes at +4"C. The cells were separated on a MiniMACS separation column according to the manufacturer's recommendations and theCDlO' and CDlO- fractions were collected. The negative cell fraction was incubated with RPE conjugated anti-CDIY MoAb, washed, and labeled with anti-isotypic MACS magnetic microbeads as described above. The cells were separated on a MiniMACS separation column and the CD19' and CD I9 fractions were collected. All cell fractions were analyzedin a fluorescence-activated cell sorter (Becton Dickinson FACSort) with the software Lysis II (Becton Dickinson). The number of CDIO', CD19' cells in the preB cell enrichedsample was 10 timeshigher than theamount of CDIO-, CD19' cells, and the ratio was reversed in the sample enriched formatureBcells.Thelymphocytefractionsenrichedfor pre-B (CDIO ') and mature B (CDl9+) lymphocytes were frozen as cellpellets at -70°C until genomic DNA preparationswere performed. Prepuration of genomic DNA. DNApreparationswere performed as described by Miller al.'" et Briefly, the cells were dissolved in a TRIS-EDTA buffer and lysed with proteinaseK and I % sodium dodecyl sulfate (SDS) in 37°C overnight. Proteins were precipitated with saturated NaCl andchromosomalDNA was recoveredfrom the aqueous phase by ethanol precipitation, and dissolved in 1 X TE (TE: I O mmol/L TRIS-HCI pH 7.6, 1 mmolL EDTA pH 8.0). The concentration and purity of the preparations was determined by measuring the absorbance at 260 and 280 nm (DU-62 Spectrophotometer: Beckman Instruments Inc. Fullerton, CA). PCK u r d PCR primers. PCR was performed in atotalvolume of SO pL AmpliTaq I X PCR buffer containing0.5 to 1 .O pg template DNA, 0.8 pmollL of the VH6 primer. 0.53 pmol/L of the JH1,2,4.5 primer.0.13pmol/L of theJJprimer, 0.13 pmol/L of theJl,6 primer, I O % glycerol, and 2.5 UAmpliTaq(PerkinElmer.Roche Molecular Systems Inc. Branchburg, NJ). The PCR was performed using a thermal cycler (Techne Ltd, Cambridge, UK) with a cycling program as follows: preheating 1 minute at 94°C; I minute at 94°C. I minute at 69"C, and 0.5 minute at 72°C for 15 cycles; I minute at 94°C. I minute at 65"C,and0.5minute at 72°Cfor 25 cycles, and finally an elongation step of 5.5 minutes at 72°C. The following PCR primerswereused: V,,6: S' CCTCTCACTCACCTGTGCCA S' AGGAGACGGTGACCAGGGT 3': JH3: S' 3': JH1,2,4.5: GAAGAGACGGTGACCATTGT 3': JH6: 5' AGGAGACGGTGACCGTGGT 3'. Cloning und sequencing. TheamplifiedPCRproductswere subcloned into the pT7-Blue(R) vector according to instructions in the pT7 Blue T-vector Kit (Novagen,Madison,WI)except that electroporation(2.5 kV, 25 mF. and200 $2. onaGenePulser; BioRad,Richmond, CA) was used fortransformation.Doublestranded DNA sequencing was performed with a Sequenase Version 2.0DNASequencing Kit asrecommended by themanufacturer (United States Biochemical, Cleveland,OH). The sequence reactions were separated on a denaturing6% acrylamide gel and detected with autoradiography. Seqrrence unalysis. Sequencingdatawereanalyzed with GCG package(HGMPResourceCentre,HarrowMiddx,UK)software Fasta and Lasergene (DNASTAR, Inc. Madison, WI). For identifi- From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 2797 lg GENE USAGE DURING RECONSTITUTION AFTER BMT Table l . Serum Ig Levels and Cellular Content in Peripheral Blood of Patient 1 and Patient 2 6 wk After 4 wk After BMT BMT 1 Total WBC 2 1 2 1 2 1 4.6 2.3 0.17 7 4.2 1.6 38 4.8 1.0 21 6.7 0.49 7 - - 6.5 0.59 9 0.5 9.4 1.0 - - - - Lymphocytes % Lymphocytes 4.1* 1.7 41 -* IgM IgG IgA 0.4t 7.0 1.5 0.3 11.4 1.5 7-8 wk After BMT BMT BMT Before BMT - - 2 10 wk After BMT 3 mo After 6 rno After 1 1 2 1 2 Normal Range 3.1 0.78 25 0.6 4.7 3.8 0.49 13 6.4 0.85 13 4.1 1.3 32 3-10 1.0-4.0 20-40 0.2 0.4-3.1 2 2.8 4.8 5.2 0.73 0.32 1.2 26 7 23 - 0.9 0.6 0.4 0.3-2.0 " - - 0.2 " 6.7.0 5 0.5 6.1 0.7 7-15 * x i 0 9 cells/L. t g/L. Not determined. * cation of D and JH genes comparisonwasmadewithpublished germline genes8.9, 31-36 The criteria for identification of a D gene segment was set to be 9-bp overlap with maximum l-bp mismatch or 10-bpoverlapwithmaximum 2-bp mismatch.Themismatches werenot allowed tohaveabiastowardtheend of theidentified fragment so thatthestretch of homology outsidetheunmatching regionwasshorterthanthemismatchstretch.If two possible D genes overlapped,the one withthe longest stretch of homology outside the overlapping region was identified or, if this stretch was of similar length for the two D genes, both genes were identified. Lymphoprep. Mononuclear cells Staining with mouse a CD10 m A b ( I g G d and MACS rat a mIg% microbeads. I Fractionation over column. "Acs I l Negative fraction; Positive cells) (pre-B l CDlO- cells '-l"--- * Staining with mouse a CD19 mAb (IgG1) and MACS rat a mIgG1 microbe ad^. I Positive fraction; CD19+ cells (mature B cells) Practionationover fraction; CD19- cells Fig 1. Strategy for enrichment of pre-B and mature B lymphocytes from the B M sample. The D gene readingframesweredefined as thefirst, second, and third frame starting from thefirst nucleotide in the germlinesequence andafunctionalrearrangement is statedwhenthere is an open reading frame through the CDR3. RESULTS Reconstitution of lymphocytes and soluble Ig afer BMT. Both patients 1 and 2 had normal white blood cell and absolute lymphocyte counts before BMT (Table 1). After the post BMT nadir, the white blood cell count increased to 2.3 X lo9 and 4.2 X lo9 cells/L, respectively, at 4 weeks and was normal 6 weeks post BMT. Six months after BMT the lymphocyte count was less than half of normal values for patient 1. Serum IgM and IgG levels were somewhat low before, as well as after, BMT for patient l and varied between 0.4 to 0.6 g/L and 4.7 to 9.4 g / L , respectively (Table 1). Patient 2 had also rather low serum levels of IgM and IgG levels varied between 6.5 and 11.4 g / L . Before BMT the serum IgA level was well within the normal range for both patients. After transplantation the IgA level decreased. Six months post BMT it was within the normal range for patient 1 but still low for patient 2. Nucleotide sequence analysis of V&-containing rearrangements. In patient 1 a total of 161 individual clones were analyzed and the complete sequences of the CDR3 regions are presented in Fig 2. The average CDR3 length is similar among the different samples (Table 2) except for the BM-derived pre-B cell population where the CDR3 length is 19.2 5 8.1 tripletcodons (P = .OS). All sequenced clones have N nucleotide additions either in the VH6-Djunction, D-JH junction, or in both. P nucleotides, as defined by Lafaille et al,I7 can be identified in some clones with similar frequency in D-JH junctions as in VH6D junctions (Fig 2). In patient 2 a total of 122 individual clones were sequenced and the CDR3 regions were analyzed for functionalrearrangements(data not shown). None of the rearrangement events identified in patient 2 were found in patient 1, and vice versa. The frequency of functional VH6-containing rearrangements in the two donors is 29% to 50% in the BM samples and 39% in the PBL sample. In PBL obtained from patient 1 before BMT 67% of the sequenced clones represent a functional rearrangement, and 4 weeks post BMT, taking both patients together, 75% of the rearrangements are func- From www.bloodjournal.org by guest on December 3, 2014. For personal use only. AND 2798 VI4 NandP D gene NASMAN N andP LUNDKVIST F/NF J, gene Donor:BM DE-B cells 3. 10. 11. 13. GGAGGG G GGGT GTCOGG GATCOG WGGACGCCC DATA AGTC A G m G A 19. 17. 5. 6. GGCATT TTCCTCTCGAG 7. m G lA ? A .G c CGGAA CGCCTTTA QA 8. 12. 14. 16. GGGACIVLCCCG TCCCCGAGATA DACTAC'IciGGccC ATTACTACTACTACTACGGTA~CGTCTOOOOCC CTACTACTACTACAlGGACGE'FOOOOCA TOACTACTDGOGCC AGA CGGCGCTMT 18. TGCAAGA 19. 20. 21. TGCA 112). 3. 6. 7. 9. 10. 19. 4. 9. 8. 11. 14. 16. 17. 18. 20. 12131 (JH6b) NF CCTCC CGATu%GcDGc GGGACAACCCG TGGGG AcGG TOCAROR GGA GAATTCCCTAACCC TCAGGGAGAA 3 3 GA 3 C GATC G 3 2 1 1 m 2 CCOOOC TAAGAGGGTACGCG TTAAGCGGG 1 3 1 1 1 3 3 G A m GATA TCACCGTC TGC 00AOCCCCCGlQ GA 2 1 3 m 3. 4. 17. 18, 19. GTC C GAGGC 3 1 CAOWLCGA AOOOAT GAGCl'ACG (JH6b) F (JH6b) F (JH4bl F IJH4bl F AGGGAG lJH4bl (JH4bl (JHSbI (JH4bl lJH4bl IJH4b) NF NF TIAGTCGCGTCA (JH6b) (JH4al (JH5b) (JHPbI lJH6b) lJH6b) NF NF NF NF NF NF C (JHSbl NF GT 23. 24. 5. GATCACTACC 1 3 2 1 1 1 3 C AGGACCACCAXAT GGA 2 Ac G A m T c G G 1 1 1 1 aAGGGC 2 AA l GG 6. 7. 8. 9. 10. 11. 12. 13. 14. 20. C'IGGTACCCAGGGT TOT GACCAGGCCWC GAT G CGcGA'IGGTTK CGAOC GGGTGG E F F NF (JH5bl NF Fig 2. The complete CDR3 region of all sequenced clones in donorlpatient pair 1. Each sequence starts with the end of FR3of the VH6 gene. The number in frontof each sequence indicates clone number and,if several clones had identical sequences, the number of repeatsis given in parentheses after the clone number. Identified D andJH genes ara specified in parentheses andthe D gene reading frameis given in front of each D gene. Abbreviations and symbols: ND, not determined; boldface letter, P nucleotides; *, deletion compared with germline sequence; underlined letter, insertion compared with germline sequence; lowercase letter, nucleotide differing from the germline sequence; -C, D gene rearrangedin reversed orientation. tional (Fig 2). At later time points the frequency decreases, but still 6 months post BMT 56% of the clones contain functional rearrangements. In general, the number of mutations in the VH6gene are low (Table 3) and the pattern of base substitutions indicate that they are of the intrinsic type."In the CDRs the BMderived mature B cells have a replacement mutational frequency of 1.5 mutations per 100 sequenced bp, resulting in an €US (replacement to silent mutations) ratio of 7.9. Four and 6 weeks post BMT the R/S ratios are 12.0 and 7.0, From www.bloodjournal.org by guest on December 3, 2014. For personal use only. lg GENEUSAGE 2799 DURING RECONSTITUTION AFTER BMT Patient PBL 2614). XULA 33. TGCAAGA 13121. T a m GAC TCAAOTrCT 1113). Toc C 3131. TGC 5 0 ) . 2 2 1 - lDIR2-Cl lD3) (DIR1-Cl (ND) 2 (DIR1) 3 lDIR2-C) 2 (DXPIJ 3 (DIRZ) 3 IDxP4) noopac-ta: 0000tcmCcGG GAMcI\cG GAGFXCAT COOOO CCCtCCUOOtGcCCCC C GTATT" CTAC Slx Weeks 2 lDIR2-Cl 1171. TGCAAGA 2111). TGC OACGCCT C 2 IDIR1) 2815). TGCAAGA GATOOO 3 lDIR2-C) 3 lW1) Seven weeks after BMT 15. 19. l(4). 3. 4. 1717). XULA 612). 912). Toc T G T m GATCKC TGCAAG TGCAAGA TGCAAG T GGCCTCT GTCTOOGMTTGG 'ECAAGA AUA - (ND) 1 2 3 2 3 (DK4-C) (DHO52) (DLR1) lDLR1) lD2-C) CCGAWLGCC 3 lDK4-Cl TGCAAGA 11. TGCAAGA 1512). 22. TGUVV;A 319). TOCAAG 2 1 4 ) . TGCAAGA TGCAA 4. GATA 2 1 1 3 2 12. TGCAAGA GGTCCCATAAT 312). TGCAAGA AA 1. TGCAAGA TGCAAGA GATAGGG GAGTCwryiAIL 3 (DXPI) 2 (m4) 1 (DZl/lO) 10. 11. TGCAAGA GATCCG (NDI T o c m - GCGCGC 6. 'ECAAGA OAocOT l. ToCA 1 1 1 3 3 2 2 1 2 (DIR2-C) (D23/7-C) (DZ-C) (D3) (D3-C) 1 2 1 2 2 2 lDK41 (m4) (W4) lDK1) (DXP'1) 5. 8. TouuLo 2. 3. 6. McAAopl 9. 5(2). 1. 4. 7. E. TACCGCCGCGTTGGA GAQXG T GGGGATCCGT T OCYH30C (03) (DKl) (DXP'11 (DXP'lI lDHQ521 IND) 1 IDIRZ) 2 lDK1) 1 (Dm) - ( n u ) ouocc TGCAAGA MOG 'ECAAGA rOcM0 'ECAAGA M - T AGACAC 2 (DK4) co1TcToo(rr 3 (m11 2 (DK1) l (D21 'ECAAGA GATcGmm AG (D21/10-C) (DIR1) (DHQ521 "OA GGAA m TA (m1 :*mx:&C Fig 2. (Cont'd) respectively. At later time points after BMT the mutational frequency and R/S ratio decrease considerably. Clonal variability. To estimate the degree of diversity in the antibody repertoire we calculated the fraction of unique sequences out of all sequences obtained from each sample, ie, if all individual clones represent unique rearrangement events the clonal variability will be 100%. As shown in Fig 3, the donors have a varied repertoire of rearrangements among BM-derived pre-B and mature B cells, as well as among PBL, with a clonal variability close to or equal to 100%.The clonal variability in patient l before BMT is 43% and the variability decreases to 13% after transplantation. In patient 2 the clonal variability is as low as 4% 4 and 6 weeks post BMT. Seven to 10 weeks post BMT the variability is 20% to 40% and at 3 and 6 months the repertoire in patient 1 is almost as diverse as among the donated cells. However, the variability remains low in patient 2. Reconstitution of the VH6-containing lg repertoire in patient l appears to follow a wavelike pattern in that one of the sequences, represented in 13 of the 19 clones isolated 4 weeks after BMT, is also present in 11 of 23 clones 6 weeks after BMT (Fig 4A). In the sample obtained 7 weeks after BMT none of the rearrangements found at 4 and 6 weeks after BMT are present. However, 4 clones at 7 weeks after From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 2800 NASMAN ANDLUNDKVIST Table 2. C M 3 Length in Samples Derived From DonorlPatient Pair 1 Before and at Different Time Points After BMT I 100 - 75 - 50 - 25 - I CDR3 Length' Donor BM pre-B cells BM mature B cells PEL Patient PEL before BMT 4 wk after BMT 6 wk after BMT 7 wk after BMT 10 wk after BMT 3 mo after BMT 6 mo after BMT 19.2 f 8.1 14.2 f 3.6 13.2 2 3.6 12.0 f 6.1 12.7 t 4.0 14.7 t 2.5 13.8 ? 4.4 14.6 ? 5.4 14.8 t 8.6 13.8 t 3.9 * CDR3 length is expressed as the average number of triplet nucleotide codons between residues 93 and 102 according to the terminology of Kabat et aI5O t SD. BMT contain a rearrangement also present in 9 clones 10 weeks after BMT and in 1 clone 3 months after BMT. Two clones obtained 3 months post BMT share the same rearrangement as well as 2 clones 6 months after BMT, but these clones are not identical. None of the 8 sequences shown in Fig 4A are present in the donor-derived samples. A similar pattern is seen in patient 2, although none of the rearrangements are found at more than one time point (Fig 4B). Also in patient 2 there is no overlap between donor and recipient derived sequences. J, gene usage. In all samples from patient 1 the Cproximal J H genes, JH4, JH5, and JH6, are most frequently used (Fig 5). In pre-B cells all rearrangements containing Table 3. Mutational frequency and Replacementto Silent Mutations Ratio @?/S)in Functional Rearrangements From DonorlPatient Pair 1 CDR FR R S R/S R S RIS 0 0.24 0.24 0 0.16 0 1.5 >0.24 0 1.5 0 0 0.19 0 7.9 4.2 1.7 0.79 1.4 2.5 2.5 1.2 >0.56 >0.11 1.0 >0.23 2.0 5.3 4.7 0 0.27 0.27 0.27 1.3 0.44 0.67 0.67 Donor BM pre-B cells BM mature Bcells PBL Patient Before BMT 4 wk after BMT 6 wk after BMT 7 wk after BMT 10 wk after BMT 3 mo after BMT 6 mo after BMT 2.0 1.7 0.56 0.11 0.11 0 0 0.23 0 0.11 0 0 0 - 1.5 12.0 7.0 0 >0.27 >0.27 >0.27 Mutational frequency is calculated as total number of nucleotide changes in the V& sequences at each sample time divided by total number of sequenced bp, multiplied with 100. Replacement (R) mutations lead to amino acid substitutions, compared to VH6 germline sequence, whereas silent (SI mutations do not affect the amino acid sequence. Framework regions (FRI are located between residues 1 to 30, 36 to 49, and 66 to 92. CDRs are located between residues 31 to 35 and 50 to 65 according to the terminology of Kabat et al.50 Fig 3. Clonal variability of the different samples. Clonalvariability is calcul8t.d as percentage of different clones in total number of sequenced clones in each sample. (A) and (B) show the clonal variability fromdonorlpatient p a i o 1 and 2, respectively. J H are ~ nonfunctional. J H 1 and JH2 are used mainly in BMderived pre-Bandmature B cells and only 1 of 161 sequences express the J H gene. ~ D gene usage. In general, D gene usage varies more among pre-B cells compared with mature B lymphocytes in From www.bloodjournal.org by guest on December 3, 2014. For personal use only. lg GENE USAGE DURING RECONSTITUTION AFTER BMT -1 L c c c 4 4 c c) I * E I 2801 gene. D2119 is used by 17.6% of mature B cells from BM, but is not present in any other sample. DM1 and D21/10 represent two D genes that are rearranged inall samples from the donor and that reappear in the patient at 3 months after BMT. DHQ52 is represented in clones isolated 7 weeks, 10 weeks, and 3 months posttransplantation, but all those rearrangements are nonfunctional. D2, DNI, and DIR2 are frequently used in clones isolated from BM and PBL from the donor as well as in PBL from the recipient. All DNIcontaining rearrangements except one are nonfunctional. The DNI gene is frequently used in pre-B leukemia, adult PBL, and in the neonatal repert~ire.~"'~ The DXP gene family, Donor: BM preBcells 1-1 75% Donor: PBL 75% Donor: BM mature B cells -1 75% m I Patient: PBL before BMT Rap l 0% Four weeks after BMT 75% Fig 4. Diversification ofthe lgrepertoire. The frequency of certain abundant rearrangements at different time points before and after BMT in patient 1 (AI and patient2 (B). In (A) sequence 1 ( 0 1 represent 13 clones from 4 weeks after BMT and 11 clones from 6 weeks after BMT. Sequences 2 (01 and 3 (A1 represent 3 clones each from 4 weeks afterBMT, sequences 4 ( 0 1 and 5 (W) represent 7 and 5 clones, respectively, from 6 weeks after BMT. Sequence 6 ( + l represent 4 clones from 7 weeks, 9 clones from 10 weeks, and 1 clone from 3 months afterBMT. Sequence 7 (01 represent 7 clones from 7 weeks and sequence 8 (AI 4 clones from 10 weeks afterBMT. In (B] sequence 1 (01represent 24 clones from 4 weeks after BMT, sequence 2 (01 23 clones from 6 weeks after BMT, and sequence 3 (A1 represent 7 clones from 8 weeks after BMT. Sequence 4 (0) and 5 (W) represent 8 clones each from 3 months after BMT and sequence 6 ( 1 represent 5 clones from thesame timepoint. I after weeks Seven BMT S i x weeks after BMT 75% Ten weeks after BMT 50% 1 + months after Three BM and peripheral blood (Fig 2). Before BMT, patient 1 uses only three different D gene segments in the VH6-containing rearrangements. With time after BMT the D gene usage becomes more diverse and is comparable with that of the donor cells 3 months post BMT. Twenty-two different germline D gene segments were represented in the samples from patient 1. Seven of those were only used in BM-derived cells and out of those Dl, DLR5, DXPI/D5, and DAI/DA4 were found only in pre-B cells. D4 is only used in the BM samples where 23.5% of the preB cells and 17.6% of the mature B lymphocytes use this BMT S i xBMT months after 1-1 75% l RI 5n%1 A 4l"J 25% 0% Fig 5. JHgene usage in VH6containing rearrangements in donor/ patient pairl.JH gene usage is expressed as percentage of the number of unique rearrangements in each sample. Functional rearrangements are represented by shaded bars (RI] and nonfunctional rearrangements by open bars (01. From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 2802 NASMANAND which has been reported to be the most frequently used D gene family in the adult PBL repertoire, is also repre~ented.’*.~~ DIR genes can be identified in 2 rearrangements before BMT, 2 rearrangements 4 weeks after BMT, and2 rearrangements 6 weeks post BMT. This high frequency of DIR gene family utilization could be due to either the low variability at 4 and 6 weeks post BMT (Fig 3A) or a selection of Bcell clones using the DIR gene in combination with VH6.In a recent report, Moore and Meekw could not demonstrate significant rearrangement to the DIR recombination signal sequences, and they suggest that these GC-rich sequences may be explained by N nucleotide additions rather than DIR utilization. We do not think this can explain our results because DIR sequences are the only D genes possible to identify in the more than 30-bp long CDR3 region isolated at 4 and 6 weeks post BMT, even though the match with reported germline sequences is not absolute. There are some clones that contain more than one D gene (Fig 2). In the pre-B cell population S of 17 clones have this type of rearrangement. Two of these clones are functionally rearrangedand three clones are nonfunctional. Four to 6 weeks after transplantation three unique rearrangements contain D-D fusions, which all are functional. D-D rearrangements are not common among the other samples and where they are present the majority are nonfunctional. D gene reading frame (RF). The junctional regions of the functional BM-derived VH6-D-JHrearrangements from patient 1 displayed a bias against D genes translated in RFl (Fig 2). In pre-B cells none of the D genes were in RF1, but among mature B cells RFI was found in 28.6% of the population. This was accompanied by a decrease in RF2 utilization in mature cells. Interestingly, this pattern is indicated also in the early posttransplantation period. Four and 6 weeks post BMT none of the rearrangements use RFI, but during reconstitution of the lymphocyte repertoire the frequency of D genes in RFI increases. Because of the limited number of unique rearrangements early after BMT, the differential utilization of D gene RFs cannot be statistically confirmed. In adult PBL, cord blood, and fetal repertoires all three D gene reading frames are usedin similar frequency, but independently of VH gene family usage, certain D genes preferentially use a specific RF.”.”Varade et a14’ found that 58% of the VH6-containing rearrangements in spleen from young individuals use RF1. DISCUSSION The well-documented overrepresentation of the v ~ gene 6 in the fetal Ig repertoire2’.22as well as in the post BMT s i t ~ a t i o n ~led ~.~ us’to perform a more detailed analysis of VH6-containing rearrangements before and at several time points after BMT. Our results show that the repertoire 4 to 10 weeks post BMT displays a limited set of rearrangements (Fig 3). This is in contrast to results derived from the BM donor samples and 3 to 6 months after BMT where there is a higher degree of variability. The high frequency of functional rearrangements and the high R/S ratios among clones isolated 4 and 6 weeks post BMT (Fig 2 and Table 3) indicate LUNDKVIST that there is a positive selection of VH6-expressing B lymphocytes during early reconstitution of the immune system after BMT. Another similarity between development of the fetal and the post BMT B-cell repertoire is the frequent use of the DN and DK gene families (Fig 2).19 In contrast, the DHQ52 gene andthe JH3 gene frequently used in the fetal repert0ire21.24.3Y are only sparsely rearranged in our material. Instead, JH gene usage in all our samples is biased toward the C-proximal genes, JH4, JHS, and JH6,(Fig 5 ) , which is the case also in both cord blood and adult peripheral blood repertoires,31.39,42.4~ Yamada et al” have described a DNl variant gene, with a CA to TG substitution, that could represent either a polymorphism or a new gene in the DN family. This gene has also been described by Mortari et a14’ and they proposed the tentative name DN2. We have also identified this variant of DNl among our samples, but since the DN 1 variant gene always is found in association with the JH6c allele and the DN1 gene in association with the JH6b allele (Fig 2), we believe that this variant represents a polymorphism rather than a new gene in the DN family. The CDR3 lengths, as shown in Table 2, do not vary much between the different sample points and are quite similar to the 12.3 t- 4.0 triplet nucleotide codons amongfetal C, transcripts reported by Mortari et A deviation is seen in the BM pre-B cell sample where the CDR3 length is 19.2 5 8.1 codons (P = .05). This long CDR3 region is partly due to frequent N nucleotide additions in both the VH6-Dand the D-JH junctions and partly torearrangement of multiple D genes. The possibility to circumvent the 12/23-bp spacer recombination rule4’maylead toincreased combinatorial diversity by D-D gene fusion. This type of rearrangement. originally suggested by Kurosawa and Tonegawa,Is is most frequent in the BM pre-B cell sample. The presence of DD fusions has previously been reported in the fetal, cord blood, and adult Ig repertoire^^'.^^.^ and evidence for the occurrence of D-D fusions as well as inverted D gene rearrangements has been p r e ~ e n t e d . ~ ” . ~ ~ Interestingly, some clones contain more than two D gene segments, for example in clone 16 and clone 18 from BM pre-B cells we can identify four D genes in each rearrangement (Fig 2). Furthermore, some clones contain more than two P nucleotides. In the suggested mechanism for P nucleotide addition a maximum of two nucleotides are added.” Our results maybe due to N nucleotides in a palindromic sequence or represent a new mechanism for Ig gene repertoire diversification. To exclude the possibility that the low variability found 4 and 6 weeks post BMT in patient 1 (Fig 3A) was caused by a bias in the PCR amplification process, we performed a second PCR on the sample material from 6 weeks post BMT followed by nucleotide sequence analysis. In Fig 2, 7 clones represented by clone 1 and 3 of the 8 clones represented by clone 2 origin from the first PCR, whereas the remaining clones were isolated in the second experiment. These 23 clones only represent three unique rearrangement events, one of which is represented in clones originating from both PCR amplifications. These results support the notion that the low From www.bloodjournal.org by guest on December 3, 2014. For personal use only. lg GENEUSAGE DURING RECONSTITUTION AFTER BM1 variability is the result of a restricted repertoire in vivo, although we cannot formally exclude the presence of other, nonamplified, rearrangements. It could be argued that the low clonal variability observed at 4 and 6 weeks post BMT in patient 1 was caused by a contaminating sequence that would be preferentially amplified when the number of B cells is verylow. However, these sequences have never previously been isolated in our laboratory and, furthermore, none of the other sequences present at 4 (clone 3 and 5) or 6 (clone 1 and 28) weeks post BMT are found at more than one time point. Therefore, we believe that the rearrangement isolated both 4 and 6 weeks post BMT represents an important clone in the early reconstitution of the Ig repertoire. To further confirm thepattern of oligoclonal diversification of the VH6containing Ig repertoire we analyzed a second BM recipient. In patient 2 the clonal variability was even lower at 4 and 6 weeks after BMT (Fig 3B) because only one unique rearrangement was found at each time point. The limited diversity of the antibody repertoire demonstrated by the low clonal variability early after transplantation (Fig 3) is in concordance with the low Ig concentrations in serum, the reduced numbers of peripheral lymphocytes and impaired polyclonai as well as specific immune responses found in BMT patients (Table l).’ Interestingly, the diversification of the repertoire is obviously not just simple additions of new clones to the existing pool of specificities. Instead it looks more like a dynamic system of appearance and disappearance of consecutive clones (Fig 4A and B). To our knowledge this type of wavelike diversification of the Ig repertoire has not been described, though there is evidence for a sequential rearrangement of human T-cell receptor V, and V+ genes during ~ntogeny.~’ It is not likely that these consecutive clones are derived from activated donor B cells or plasma cells transferred with the BM graft because we do not detect any of these rearrangements in the BM or PBL samples from the donor. It has been shown that secretory Ig levels in saliva peak 2 to 3 weeks after BMT and that these Igs are derived from activated cells from the BM donor.48 The Ig levels thereafter gradually decrease and remain low for around 3 months. When the levels increase again the Igs originate from the reconstituted B-cell compartment. The same pattern was found for Ig levels in serum even though the origin of the antibodies was not ~ h o w n . 4These ~ findings further support our conclusion that the pattern ofIg gene expression during reconstitution of the B-cell repertoire after BMT is a property of the new immune system developing from graft-derived hematopoetic precursors. ACKNOWLEDGMENT We are grateful to Dr 0. Ringdkn, L. Markling, M. Remberger, and C. Tammik for providing us with sample material. We also thank Drs D. Holmberg, S . Pettersson, and 0. Ringden for critical reading of the manuscript. REFERENCES 1. Storek J, Saxon A: Reconstitution of B cell immunity following bone marrow transplantation. Bone Marrow Transplant 9:395, 1992 2. Storek J, Ferrara S, Ku N, Giorgi JV, Champlin RE, Saxon A: B cell reconstitution after human bone marrow transplantation: Recapitulation of ontogeny? Bone Marrow Transplant 12:387, 1993 2803 3. Velardi A, Cucciaioni S , Terenzi A, Quinti I, Aversa F, Grossi CE, Grignani F, Martelli M F Acquisition of Ig isotype diversity after bone marrow transplantation in adults. A recapitulation of normal B cell ontogeny. J Immunol 141:815, 1988 4. Small TN, Keever CA, Weiner-Fedus S, Heller G, O’Reilly RJ, Flomenberg N: B-cell differentiation following autologous, conventional, or T-cell depleted bone marrow transplantation: A recapitulation of normal B-cell ontogeny. Blood 76:1647, 1990 5 . Kagan JM, Champlin RE, Saxon A: B-cell dysfunction following human bone marrow transplantation: Functional-phenotypic dissociation in the early posttransplant period. Blood 74:777, 1989 6. Witherspoon R P , Storb R, Ochs HD, Fluornoy N. Kopecky KJ, Sullivan KM, Deeg JH, Sosa R, Noel DR, Atkinson K, Thomas ED: Recovery of antibody production in human allogeneic marrow graft recipients: Influence of time posttransplantation, the presence or absence of chronic graft-versus-host disease, and antithymocyte globulin treatment. Blood 58:360, 1981 7. Korsmeyer SJ, Elfenbein GJ, Goldman CK, Marshall SL, Santos GW, Waldmann TA: B cell, helper T cell, and suppressor T cell abnormalities contribute to disordered immunoglobulin synthesis in patients following bonemarrow transplantation. Transplantation 33:184, 1982 8. Ichihara Y, Matsuoka H, Kurosawa Y: Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J 7:4141. l988 9. Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P: Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell 27:583, 1981 10. Tonegawa S: Somatic generation of antibody diversity. Nature 302575, 1983 1l . Yancopoulos GD, Alt FW: Regulation of the assembly and expression of variable region genes. Annu Rev Immunol4:339, 1986 12. Kodaira M, Kinashi T, Umemura I, Matsuda F, Noma T, Ono Y, Honjo T: Organization and evolution of variable region genes of the human immunoglobulin heavy chain. J Molec Biol 190529, 1986 13. Berman JE, Mellis SJ, Pollock R, Smith CL, Suh H, Heinke B, Kowal C, Surti U, Cantor CR, Alt FW: Content and organization of the human Ig VH locus: definition of three new VH families and linkage to the Ig CH locus. EMBO J 7:727, 1988 14. Buluwela L, Rabbits TH: A VH gene is located within 95 kb of the human immunoglobulin heavy chain constant region genes. Eur J Immun 18:1843, 1988 15. Kurosawa Y, Tonegawa S : Organization, structure, and assembly of immunoglobulin heavy chain diversity DNA segments. J Exp Med 155:201, 1982 16. Alt F W ,Baltimore D: Joining of immunoglobulin heavy chain gene segments: Implications from a chromosome with evidence of three D-JH fusions. h o c Natl Acad Sci USA 79:4118, 1982 17. Lafaille JJ, De Cloux A, Bonneville M, Takagaki Y, Tonegawa S : Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell 592359, 1989 18. French DL, Laskov R, Scharff MD: The role of somatic hypermutation in the generation of antibody diversity. Science 244:1152, 1989 19. Kirkham PM, Mortari F, Newton JA, Schroeder HW Jr: Immunoglobulin V, clan and family identity predicts variable domain structure and may influence antigen binding. EMBO J 11:603, 1992 20. Cook GP, Tomlinson IM, Walter G , Riethman H, Carter NP. Buluwela L, Winter G , Rabbits TH: A map of the human immunoglobulin VHlocus completed by analysis of the telomeric region of chromosome 14q. Nat Genet 7:162, 1994 21. Cuisinier AM, Guigou V, Boubli L, Fougereau M, Tonnelle From www.bloodjournal.org by guest on December 3, 2014. For personal use only. 2804 C: Preferential expression of VHS and VH6 immunoglobulin genes in early human B-cell ontogeny. Scand J Immunol 30:493, 1989 22. Berman JE, Nickerson KG, Pollock RR, Barth JE, Schuurman RK, Knowles DM, Chess L, Alt F W : VH gene usage in humans: biased usage of the VH6 gene in immature B lymphoid cells. E J Immunol 21:1311, 1991 23. Schroeder HW Jr, Hillson JL, Perlmutter RM: Early restriction of the human antibody repertoire. Science 238:791, 1987 24. Schroeder HW Jr, Wang JY: Preferential utilization of conserved immunoglobulin heavy chain variable gene segments during human fetal life. Proc Natl Acad Sci USA 87:6146, 1990 25. Guigou V, Cuisnier A-M, Tonelle C, Moinier D, Fougereau M, Fumoux F: Human immunoglobulin VH and Vk repertoire revealed by in situ hybridisation. Mol Immunol 27:935, 1990 26. Zouali M, Theze J: Probing VH gene-family utilization in human peripheral B cells byin situ hybridization. J Immunology 146:2855, 1991 27. Fumoux F, Guigou V, Blaise D, Maraninchi D, Fougereau M, Schiff C: Reconstitution of human immunoglobulin VH repertoire after bone marrow transplantation mimics B-cell ontogeny. Blood 81:3153, 1993 28. Storek J, King L, Ferrara S, Marcelo D, Saxon A, Braun J: Abundance of a restricted fetal B cell repertoire in marrow transplant recipients. Bone Marrow Transplant 14:783, 1994 29. Ringden 0, Pihlstedt P, Markling L, Aschan J, B&yd I, Ljungman P, Lonnqvist B, Tollemar J, Janossy G, Sundberg B: Prevention of graft-versus-host disease with T cell depletion or cyclosporin and methotrexate. A randomized trial in adult leukemic marrow recipients. Bone Marrow Transplant 7:221, 1991 30. Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1988, 1988 31. Yamada M, Wasserman R, Reichard BA, Shane S, Caton AJ, Rovera G: Preferential utilization of specific immunoglobulin heavy chain diversity and joining segments in adult human peripheral blood B lymphocytes. J Exp Med 173:395, 1991 32. Siebenlist U, Ravtech JV, Korsmeyer S, Waldmann T, Leder P: Human immunoglobulin D segments encoded in tandem multigenic families. Nature 294:63 I , 1981 33. Bakhshi A, Wright JJ, Graninger W, Seto M, Owens J, Cossman J, Jensen JP, Goldman P, Korsmeyer SJ: Mechanism of the t(14; 18) chromosomal translocation: structural analysis of both derivative 14and 18 reciprocal partners. ProcNatlAcadSciUSA 84:2396, 1987 34. Matsuda F, Shin EK, Hirabayashi Y, Nagaoka H, Yoshida MC, Zong SQ, Honjo T: Organization of variable region segments of the human immunoglobulin heavy chain: duplication of the D5 cluster within the locus and interchromosomal translocation of variable region segments. EMBO J 8:2501, 1990 35. Zong SQ. Nakai S, Matsuda F, Lee KH, Honjo T: Human immunoglobulin D segments: Isolation of a new D segment and polymorphic deletion of the D1 segment. Immunol Lett 17:329, 1988 NASMANAND LUNDKVIST 36. Buluwela L, Albertson DG, Shemngton P, Rabbitts PH, Spurr N, Rabbitts TH: The useof chromosomal translocations to study human immunoglobulin gene organization: Mapping DH segments within 35 kb of the C mu gene and identification of a new DH locus. EMBO J 7:2003, 1988 37. Betz AG, Neuberger MS, Milstein C: Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunol Today 14:405, 1993 38. Wasserman R, Ito Y, Galili N. Yamada M, Reichard BA, Shane S, Lange B, Rovera G: The pattern of joining (JH) gene usage in the human IgH chain is established predominantly at the B precursor cell stage. J Immunol 14951 1, 1992 39. Sanz I: Multiple mechanisms participate in the generation of' diversity ofhuman H chain CDR3 regions. J Immunol 147: 1720. 1991 40. Moore BB, Meek K: Recombination potential of the human DIR elements. J Immunol 154:2175, 1995 41. Varade WS, Marin E, Kittelberger AM, Insel RA: Use of the most JH-proximal humanIg H chain V region gene, VH6, in the expressed immune repertoire. J Immunol 150:4985, 1993 42. Mortari F, Newton JA, Wang JY, Schroeder H Jr: The human cord blood antibody repertoire. Frequent usage ofthe VH7 gene family. Eur J Immunol 22:241, 1992 43. Huang C, Stewart AK, Schwartz RS, Stollar BD: Immunoglobulin heavy chain gene expression in peripheral blood lymphocytes. J Clin Invest 89: 1331, 1992 44. Mortari F, Wang JY, Schroeder HW Jr: Human cord blood antibody repertoire. Mixed population of VH gene segments and CDR3 distribution in the expressed C alpha and C gamma repertoires. J Immunol 150: 1348, 1993 45. Rathbun GA, Tucker PW: Conservation of sequences necessary for V gene recombination, in Kelsoe G, Schultz D (eds): Evolution of the Immune Response. New York, NY, Academic, 1986, p 75 46. Meek KD, Hasemann CA, Capra ID: Novel rearrangements at the immunoglobulin D locus. Inversions and fusions add to IgH somatic diversity. J Exp Med 17039, 1989 47. Krangel MS, Yssel H, Brocklehurst C, Spits H: A distinct wave of human T cell receptor gammddelta lymphocytes in the early fetal thymus: Evidence for controlled gene rearrangement and cytokine production. J Exp Med 1725347, 1990 48. Chaushu S, Chaushu G, Garfunkel AA, Slavin S, Or R, Yefenof E: Salivary immunoglobulins in recipients of bonemarrow grafts. I. A longitudinal follow-up. Bone Marrow Transplant 14:871. 1994 49. Brenner MK, Wimperis JZ, Reittie JE, Patterson J , Asherson GL, Hoffbrand AV, Prentice HG: Recovery of immunoglobulin isotypes following T-cell depleted allogeneic bone marrow transplantation. Br J Haematol 64:125, 1986 SO. Kabat EA, Wu T T , PerryHM, Gottesman KS, Foeller C: Sequences of proteins of immunological interest. Bethseda, MD, NlH Publication No. 91-3242, 1991
© Copyright 2026