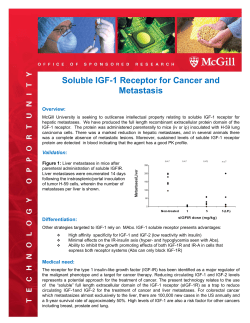
Chapter 1 General overview.
Chapter 1 General overview. 1.1. General Introduction Ever since the first protein structures were solved by means of X-ray crystal diffraction in the late 1950’s and the creation of the protein databank (PDB) in 1971 a great potential emerged for the research of protein functionality, novel drug design as well as the structure prediction of unsolved proteins. In the past two decades a substantial amount of progress has been made in terms of protein structure discovery. The solution of protein structures has become substantially easier with the development of new techniques such as Nuclear Magnetic Resonance spectroscopy and the evolution of X-ray analysis which has resulted in excess of 30,000 deposited entries of structures in the PDB. Nevertheless the three-dimensional structure of many important proteins remains unsolved. Furthermore, with the recent complete mapping of the human genome, the number of proteins sequences that lack a corresponding tertiary or quaternary structure has largely increased. This substantial increase in novel putative targets for drugs needs to be addressed efficiently and this requires new approaches and inventive tools. At present, two alternative, yet complementary, techniques for drug testing of protein targets are engaged: experimental high-throughput screening (eHTS) of large compound libraries and virtual High Throughput screening (vHTS) of ligand databases increasingly provided by combinatorial chemistry and computational methods. There are significant disadvantages in the experimental method such as the increased cost and time limitations. On the other hand virtual High Throughput Screening significantly reduces both the cost and time needed to virtually trial hundreds of thousands or even millions of compounds from combinatorial chemistry libraries on to suitable targets for virtual screening and de novo design. The currently largest vHTS project in the world, the ScreenSaver LifeSaver Project is discussed in Chapter 3. A current estimation of the total number of human genes suggests that there may be approximately 80,000 human genes. If the effect of posttranslational modification is taken into account, it has been inferred that in the Homo sapiens there may be as many as 120,000 different proteins. This figure in comparison to the number of complete protein structures currently solved (much less than the 30,000 in the PDB as there is a significant of overlap and incomplete or modelled structures) also demonstrates the need for novel efficient protein modelling and model evaluation techniques. Certain families of proteins have the ability to produce crystals more readily than others. It is commonly known that soluble proteins are generally easier to crystallise than membrane bound or transmembrane proteins. The G-coupled protein receptors, a 7 helix transmembrane family of proteins is one of these. Currently there is only one member of the GPCR family with a solved structure and this is bovine rhodopsin. In this thesis the modelling of a crucial member of this family, the β2 adrenergic receptor is discussed, both in vacuo (chapter 5) and in a simulated membrane environment (chapter 7) as well as its evaluation in terms of enrichment, hit rate and yield using vHTS. Furthermore the modelling and evaluation of another two receptors of this family, the bradykinin and chemokine receptors are discussed in chapter 8. It is commonly accepted that there both because of hardware and software limitations, vHTS simulations may be subject to a number of false positive or false negative hits. In chapter 6, a significant effector in terms of GPCR related molecules, ligand promiscuity is considered and in chapter 9, a novel method for post screening result filtering method based on the naïve Bayesian method is examined. Finally, in chapter 10, using Molecular Dynamics simulations, an effort to investigate the activation of a G-protein (transducin) is discussed as well as the importance of the domain movement of different regions of the protein with regards to the signal transduction after GPCR binding. Chapter 3 The ScreenSaver LifeSaver Project 3.1 Introduction Virtual high throughput screening (vHTS), or in silico screening, is a new approach that is now attracting increasing levels of interest in the pharmaceutical industry since it has shown promise as a productive yet costeffective technology in the search for new drugs. Even thought the computational analysis of chemical databases to identify leads for a given biological target has been practiced for many years in molecular modelling groups, the availability of low-cost high-performance computing platforms has transformed the process so that increasingly complex and more accurate analyses can be performed on very large data sets of thousands, millions or even billions of compounds in compound databases. A key aim of vHTS is to reduce the number of new structures that chemists need to generate as unpromising compounds are eliminated before synthesis while the quality of structures selected for synthesis and testing is correspondingly improved. [1,2] Randomly screening large numbers of molecules against a protein target in a laboratory is one approach to drug discovery that could best be compared to looking for a needle in a haystack. On the other hand a single computer can perform all the calculations needed to test whether drug-like molecules from a large database are likely to bind to a given protein target in a fraction of the time it would take to carry out the in vivo or in vitro tests. 3.2 Screensaver Lifesaver The Screensaver Lifesaver project is based on the idea of distributed computing. The University of Berkeley pioneered distributed computing with the SETI@home (Search for Extra Terrestrial Intelligence) project (setiathome.ssl.berkeley.edu). The software distributed to the millions of space enthusiasts via the web was designed to find patterns in radio data signals received from space. So far, although it appears to be unfruitful in terms of finding alien life forms, it has been extremely successful in highlighting the potential processing power that can be exploited from idle computers. For most of the day, approximately 95% of the CPU power of an average PC is unused and could be harnessed to carry out worthwhile tasks. One such worthwhile cause is the ‘Screensaver Lifesaver project’ (http://www.chem.ox.ac.uk/curecancer.html), which currently uses the ‘idle’ CPU power of more than 3 million computers in more than 200 countries around the world; this is a collaboration between the Universities of Oxford and Essex and the commercial companies UD and Accelrys, with the University of Essex playing a major role in defining the binding sites of various cancer related target proteins. The distributed computing expertise has been provided by United Devices (www.ud.com) and companies such as Intel, Microsoft, IBM and the National Foundation for Cancer Research (NFCR) have sponsored this project. The United Devices central server is responsible for providing the screensaver to the PC users but also receives and validates the returned results. It is important to note that the application is suspended when the user interacts with their PC; the screensaver therefore only utilises free CPU cycles so as not to interfere with the PCs performance at other times. [3] The screensaver, which can be downloaded for free from the internet (www.grid.com), facilitates calculations of the binding energy of different drug like molecules (from a database of 35 million molecules) to the target proteins. While calculations on each of the molecules may take 20 minutes on a single PC, an excess of 417,000 years of CPU power (May 2005) has been harnessed from the volunteering PCs from what is currently the world’s second largest virtual supercomputer. Figure 5.1. The Screensaver as it appears when the primary docking task is executing. The left panel shows the current target protein in a CPK coloured, space filling view, the right panel shows a 3D stick diagram of the current ligand. 3.2.1 Phase I Phase I of this project called CAN-DDO started in April 2001. The software utilized in this phase was THINK (To Have Information aNd Knowledge) written by Keith Davies of Treweren Consultants. During this phase 35 billion compounds derived from suppliers catalogues and combinatorial libraries and 100 conformers for each were screened bringing the total number of drug-like ligands to a staggering 3.5 billion. Unavoidably this would include some duplicate drug-like molecules but that did not have a major computational strain overall. Then their pharmacophores where matched to the proteins pharmacophore. After the ligands were virtually docked in the target protein they were then scored to quantify the strength of the interactions with the protein and the ranking was based on the Chemscore equation described in Chapter 2. The scores when there returned the UD server which separated them to hits or 'non hits' according to their score in terms of binding energy. Ideally the number of hits should be small to minimize the amount of network traffic and so that the promising ligands can be tested in the lab. This was the case for some of the target proteins but as predicted, because of the massive size of the screening database, some other targets got thousands or even hundreds of thousands of hits. The molecules which scored lower than 1000 kJ/mol were returned as hits and the histograms (examples below) show the distribution of these crude hits as a function of the score. Scores or free energy binding predictions below zero correspond to molecules which might be binding to and inhibit the protein target. 4500 12000 4000 3500 Number of Hits Number of hits 10000 8000 6000 4000 3000 2500 2000 1500 1000 2000 500 0 0 <0 <100 <200<300 <400<500 <600<700<800<900<1000 <0 <100<200<300<400<500<600<700<800<900<1000 Binding free energy (kJ/mol) Binding free energy (kJ/mol) 20000 140000 18000 120000 14000 Number of Hits Number of hits 16000 12000 10000 8000 6000 100000 80000 60000 40000 4000 20000 2000 0 0 <0 <100 <200 <300 <400 <500 <600 <700 <800 <900 <1000 Binding free energy (kJ/mol) <0 <20 <40 <60 <80 <100 <120<140 <160<180 <200 Binding free energy (kJ/mol) Figure 5.2. Four examples of set of results returned to the United Devices server. On the top left, an ideal case scenario where the target Fibroblast Growth Factor Receptor gave rise to 58 hits with a predicted binding free energy below zero, a manageable number as these components can easily be screened in vivo. As the binding free energy increases, the number of hits increases as well as there are more not-specific binders as expected. However, this was the only target with such a reasonable number of predicted binders. On the top right, the second best target in terms of virtual screening was cyclooxygenase with 568 ligands with free energy binding predictions below zero. In the bottom, the target RAF gave rise to 6,103 and on the right, an entirely failed case scenario with protein tyrosine phosphatase 1B which gave rise to a massive number of false positive binders (127,878 energy predictions below zero). 3.2.2 Phase II Mostly due to the enormous number of hits returned for some protein targets in this phase as well as the advance and development of more elaborate docking and scoring algorithms it was decided to re-evaluate the project and screen the ligands (without the extra generated conformers from THINK) with ligandfit. 3.2.2.1 Protein Preparation The proteins 3-D coordinates were retrieved from the (PDB) [4]. A problem with this particular format is that in this database is that the proteins only have they only contain information about the proteins heavy atoms. For docking purposes, the hydrogen information is crucial therefore these are added with the hydrogen builder module from Cerius2. Other protein modification may involve removal of the proteins co-crystallised ligands, removal of lone pairs, removal of solvent that was not present in the binding site and finally removal of duplicate atoms and general editing according to the protein. 3.2.2.2 Binding Site Definition The software used for defining the active site of the protein targets was ligandfit [5], as implemented in the Cerius2 software package distributed by Accelrys. There are two approaches for defining the binding site. The first involves using the cavity search algorithm from ligandfit (a flood filling algorithm) to search for possible large cavities where ligands can bind. The second involves using the coordinates of the ligand that has co-crystallized with the protein structure as a reference for the binding site location. The single most important element in determining the binding site of the target proteins was the information gathered from the literature. Mutational studies are a very useful source for providing information with regards to key catalytic residues which need to be included. Furthermore, some of those proteins were crystallised with their natural ligands already bound. This provides excellent information about key residues of a binding site as well as the vital interactions that need to be satisfied between the ligand and the enzyme. In cases where there is no extensive data for the catalytic residues in the literature and a ligand was not co-crystallised with the protein several other techniques are used. Based on the flood filling algorithm prediction the most suitable cavities were selected and later expanded or contracted in different areas according to the spatial availability around the predetermined site, the potential accessibility for a ligand as well as the adjacent environments electrostatics and hydrophobicity. For the analysis of the protein electrostatics around the binding site the electrostatics and Brownian/Molecular Dynamics program UHBD (University of Houston Brownian Dynamics) [6] was used in combination with the electrostatics and hydrophobicity plotting modules available in the Cerius2 interface. Then the binding site was enlarged or retracted accordingly in order to encompass additional identified regions of interest. The final step was to re-dock the natural ligand (the ligand that had cocrystallized with the protein or protein domain under examination) back in the protein where it was originally extracted from in order to examine the binding sites’ behaviour. The principal behind this is that if the site was determined properly and the right parameters have been used during the docking process then the docking algorithm should be able to reproduce the same docking pose with the smallest root mean square distance (RMSD) as possible as with the example shown in figure 3 below where the natural ligand of HIV- protease 4PHV has been re-docked in the defined binding site for the protein with a very small RMSD. HIV- protease was one of the proteins in the training set of ligandfit and is therefore the program is expected to perform well in this target as shown below. Figure 5.3. The crystal structure of HIV protease (white, space filled) complete with the active site residues (grey, ball and stick) and the ligand (4PHV, gold stick diagram). The figure also shows the location of the docked ligand (green, stick). The ligandfit software docks the ligand back to its original location with a very small, almost negligible RMSD. Re-docking a protein’s natural ligand in its active site is a standard practice to establish the validity of the active site model and the parameters used in the docking procedure. 3.2.2.3 Time Saving Due to the vast screening size of this project every possible effort was taken to make the docking procedure as time efficient as possible with the smallest possible compromise in terms of accuracy. In order to achieve this, the set of parameters which would be used for the docking were put through a number of tests using established protein-ligand complexes so as to determine the most appropriate cut-off points. One of the most time consuming elements of the docking procedure is the partitioning of the binding site into sub-partitions. Even though this practice clearly increases the accuracy of the docking, it adds a great strain in terms of the amount of calculations needed per ligand and this results in the following relationship between the number of site partitions and the time needed for docking per ligand on an SGI Octane platform (as shown in figure 4 below). 30 CPU time per ligand (min) 25 20 15 10 5 0 1 2 3 4 5 6 Num ber of Partitions Figure 5.4. The relationship between the number of site partitions and time needed for docking per ligand. For this particular test the Cyclin Dependent Kinase enzyme was the target protein and the ligand docked was olomoucine. A similar trend is observed with other combinations of proteins/ligands. A steep time increase is always observed when 4 partitions are used so it was decided that the ideal number for this project was 3 partitions. The number of partitions used is actually 6 (1+2+3) Another time saving step in the processing of the binding site is the removal of flares and oddities where ligand side chains cannot fit. This saves a substantial amount of time when calculating the energy grid as well as in the actual docking procedure (I could add a picture here…). Furthermore, in the docking parameters the ligand reject number of steps is set to 400. This means that a ligand is rejected if it cannot fit the binding site after 400 steps saving unnecessary computational time when ligands would clearly not be suitable for a particular binding site (usually because they are oversized or rigid/planar ligands in complex non-cleft like binding sites. 3.2.2.4 Other ligandfit parameters - Docking Flexible fit and not rigid fit was used to dock the ligands in the binding sites. Even though allowing for flexibility drastically increases the computational time needed it also radically increases the procedure’s accuracy as most ligands are flexible with a significant number of degrees of freedom. The protein itself however remains rigid with no degree of flexibility or rotating side chain groups as seen in other programs. The 5 best scoring poses for each ligand are saved. The ligands are energy minimised in the protein (maximum number of iterations= 100). The polar torsion steps is 30 degrees. No clustering is used because even though there might be some similar hits because of the Monte Carlo nature of the docking. Each ligand is given 25000 trials. All the simulations are done in vacuo (dielectric =1), and no solvent or membrane environment are accounted for due to the serious time restraints this would cause as well as the docking software limitations. Finally the forcefield used for this experiment was the latest version of the CFF at the time (version 1.55) and the scoring functions to evaluate the docking were Ligscore 1, Ligscore 2, PLP1, PLP2, PMF and Jain therefore giving the ability of consensus scoring. These scoring functions and the CFF forcefield are described in more detail in Chapter 2. 3.2.2.5 The ligand database The database contains 35 million different small compounds; of these 1.4 million compounds came from a list of unique compounds that can be found in catalogues of commercial suppliers. This set served as a starting point. Several well characterized [7-9] combinatorial chemical libraries were added which pushed the number up to more than 1 billion molecules. The structures were filtered down to 35 million to make sure that only molecules with drug-like properties were included (e.g. Lipinski’s rules of 5). [10-12] Such properties include appropriate molecular mass (150-800 Da) and solubility. 3.2.2.6 The protein Targets The earliest anti-cancer drugs most commonly used in chemotherapies worked by inhibiting cell growth, resulting in cell toxicity which triggers apoptosis (programmed cell death). Although this is the desirable effect for the fast dividing cancer cells, unfortunately this is a non-specific method which results in a wide range of side effects, the most common of which are nausea and vomiting, hair loss, and bone-marrow depression, changes in taste, fatigue, and changes in smell perception [12]. In this virtual screening project, the target is not to stop cell growth in general but to inhibit the normal functions of oncogenic proteins. However, especially for kinases such as Raf or c-Abl tyrosine kinase, it will be fairly difficult to mine target specific inhibitors as most kinase inhibitors are directed towards the conserved ATP binding site. The key recognition features of the ATP binding site are conserved in all the 518 putative protein kinases in the human genome [82].Because the essential features of this site are conserved in all eukaryotic protein kinases, it is generally assumed that the same compound will bind in a similar manner to different protein kinases. Every effort possible was made to ensure that any distinctive features (such as non-conserved residues) of the regions in the binding site vicinity were included in our definition of the site in the effort to identify target specific drugs as small differences in the constellation of residues adjacent to the site have allowed, through chemical screening and structure-based methods, high affinity compounds to be developed that are selective for just a few kinases [16]. The function, size and properties of these proteins vary significantly. As an example we can briefly compare two of the protein targets; Ras (pdb code: 821P) and Cyclooxygenase (pdb code: 6COX). Ras has 166 residues and is a mutant of the P21 Protein where Glycine 12 is replaced by a Proline. P21 is a very important molecule as it acts as a tumour suppressor mechanism (arrests cell cycle) and its mutant form is found in many cancer patients and knock out mice. [15] Cyclooxygenase is a much larger protein (homodimer with 1100 residues). Its function is to keep the blood vessels open and flowing in areas where there is tissue damage or swelling that often occurs around certain cancerous growths. Therefore by inhibiting this target, the blood supply to the cancer cells is interrupted and cancer cells cannot survive. [18] The protein targets can be divided in several families based on their function. A number of kinases and phosphatases are included which are responsible for the regulation of very important cellular processes and molecular cascades and may facilitate the growth of cancer cells (e.g. Insulin Tyrosine Kinase, CDK2, Protein-Tyrosine-Phosphatase 1B). These enzymes which regulate the cell cycle are very often involved in the pathogenesis of cancer as they promote uncontrolled growth or help the cancerous cells to avoid programmed cell death (apoptosis). Furthermore a number of cytokines and growth factor polypeptides have been included which have been shown to act as survival factors during angiogenesis, including the basic fibroblast growth factor receptor (FGFR) and vascular endothelial growth factor (VEGF) and its receptor (VEGFR1). Basic FGF (bFGF) and VEGF are two of the cytokines that have been most widely studied, because of their ability to induce many physiological responses, including survival and tumour growth. Both in vitro and in vivo studies suggest that these mediators play a role in angiogenesis [14]. Finally other proteins include RAS which interacts with RAF and a large number of other proteins and result in cell growth activation, cyclooxygenase 2 which similarly to the Growth Factors and their receptors leads to vasodilation and therefore increased blood supply for the tumour cells and superoxide dismutase which removes the toxic superoxide from cells. The function and information on the binding site definition of each of these proteins is explained in more detail in this chapter. Table 5.1. The current protein targets of the Screensaver Lifesaver project, and the expected effect of their inhibition PDB code 821P Name RAS Cyclooxygenase Inhibition Effect Ref. Control of cancer cell growth 15 Decrease of blood supply to cancer cells leading 6COX 18 (COX-2) to their death Vascular Endothelial Prevention of development of blood vessels 1FLT Growth Factor 19 leading to decreased blood supply to cancer cells (VEGF) Inactivation of VEGF leading to same effect as 1FLT VEGF Receptor 1 19 targeting VEGF directly Superoxide Stops removal of superoxide radicals from cancer Dismutase cells leading to self oxidisation of cancer cells 1ISC 20 Insulin Tyrosine 1IR3 Reduced Cancel cell growth 21 Kinase C-ABL Tyrosine Prevent development of Leukaemia and shrinkage 1IEP 22 kinase Fibroblast Growth of certain tumours Decrease of blood supply to cancer cells leading 1AGW 23 Factor Receptor to their death Cyclin Dependant 1AQ1 Deregulation of cancer cell cycle 24 Kinase 2 (CDK-2) 1C1Y RAF Inhibition of cancer cell growth 25 1D8D Farnesyl-transferase Inhibition of cancer cell growth 26 Inhibition enables apoptosis of cancer cells 29 Protein-Tyrosine1BZH Phosphatase 1B 3.2.2.6.1 Target 1: H-RAS protein Biological significance The Ras protein is one of the most important members of a large super-family of proteins known as "low-molecular weight G-proteins” or small G-proteins. These proteins are called "G-proteins" because they bind guanine nucleotides (GTP and GDP). They are called "low-molecular weight" to distinguish them from another, distinct, family of guanine nucleotide-binding proteins, the heterotrimeric G-proteins such as transducin examined in chapter 7. The gene of this small G-protein was the first oncogene to be isolated from human cancer cells (Kolch from Raf). These relatively small proteins play a very important role in cell growth regulation and differentiation and have been implicated in the process of malignant cell transformation that leads to carcinogenesis [30]. Mutationally activated and oncogenic versions of the Ras genes were first identified in human tumors in 1982 and today we know that approximately 30% of all human tumours are believed to arise from a mutated Ras oncogene and [3133]. More specifically, point mutations in the corresponding Ras genes are found in over 90% of human pancreatic carcinomas and 50% of human colon cancers which is the third leading cause of cancer in males, fourth in females in the U.S [34]. As a biochemical consequence of those mutations the Ras protein loses its GTPase activity and therefore permanently stays in the GTPbound active state sending constantly signals into the nucleus. This in turn results in uncontrolled cell division being characteristic for cancer diseases. [15] Because of their massive impact in cell cycle regulation and proliferation, wild form Ras and the most common mutants have warranted an extensive amount of research. A number of different strategies has been deployed in different occasions in order to block its activation (which in turn leads to raf-1 activation and a cascade that ends in the cell nucleus and the activation of the AP-1 gene expression sending the signal to the cell to proliferate). There are currently 5 different anti-Ras strategies that are currently under evaluation in the clinic are pharmacologic inhibitors designed to prevent: (1) association with the plasma membrane (farnesyltransferase inhibitors as explained later in this chapter), (2) downstream signaling (Raf and MEK protein kinase inhibitors also explained in more detail when discussing Raf inhibition), (3) autocrine growth factor signaling (EGF receptor inhibitors), (4) gene expression (H-ras and c-raf-1) and (5) as we are aiming here a direct inhibition of the Ras protein by identifying potent antagonists which would block the ATP binding site of the protein therefore not allowing its activation. Protein structure and Binding site definition The three-dimensional structure of the Ras protein has been determined for both the GDP-bound form (amino acids 1-169, pdb code 4q21) [35] and a non-hydrolysable GTP-bound form (amino acids 1-166) which was the structure used for this project (pdb code 821p) solved by Scheidig et al at 1.50Å [15]. The Ras protein is encoded by a 189 codon open reading frame, and is synthesized as such in E. coli. The X-ray crystallographic studies were made from premature termination mutants of H-Ras made- in E. coli, since these crystallized more easily. An examination of the tertiary structure of this target reveals an alpha/beta fold with 6 beta-strands comprising a central beta-sheet surrounded by 5 alpha-helices (Figure 6 below). The positions that change the most in the GTP versus GDP bound forms are located close to the third (gamma) phosphate in the GTP as shown in figures 5 and 6 below. The regions are called the 'switch I', residues 30 - 38, and 'switch II', residues 60 76 [36] This particular structure used for project is one of the most common mutants of the protein where Glysine 12 is mutated to a proline (G12P). Other common mutations involving Glysine 12 are G12R and G12V but glutamine 61 is also commonly mutated (e.g. Q61H and Q61L). The binding of GTP is what activates Ras to send signals to the cell that it should divide. The MG ion is also very close and is considered part of the active site. To be in the active site the amino acids need to be within about 3.0 angstroms from the GTP molecule. If any of the amino acids in the active site are mutated, the result could change the interaction between Ras and GTP. If the mutation prevents GTP from disassociating with Ras, the cell could begin uncontrolled division [33]. The biochemical basis for the GTPase defect in oncogenic Ras has been partially revealed. Glutamine-61 of Ras appears to play a role in activating a water molecule for nucleophilic attack, and its substitution by any other amino acid abolishes both intrinsic and GAP-stimulated GTPase activity hence it was included in the binding site. The crystal structure of the RasRasGAP complex revealed that any mutation of glycine-12 or glycine-13 would position a side-chain that both displaces glutamine-61 and sterically occludes the catalytic "arginine finger" of GAP (Arg 789), resulting in a loss of intrinsic and GAP-stimulated GTPase activity [37]. A number of other residues are involved in the binding site as shown in figure 7 below. Glycine 13 and Phenylalanine 28 are involved in hydrophobic contacts with the ligand and so do partially Glycine 15 (CA), Alanine 18 (CB) and Lysine 117 (CA, CG and CE form hydrophobic contacts with the C5, C6 and C8 of the rings of ATP). The following residues included in the definition of the binding site form hydrogen bonds with ATP: Gly 15, Ser 17, Ala 18, Gly 60, Asn 116, Lys 117, Asp 119, Ala 146. Furthermore, Ser 17, Thr 35 and two oxygens from the phosphate groups of ATP interact with the Mg ion. Finally, a there is a number of water molecules which could potentially act as hydrogen bond bridges between the ATP and residues that are slightly further away from the main core of the binding site such as Glu 31 and Asp 57. However, as the water molecules have been removed from the protein their effect may be undermined in this screening. Figure 5.5. The three dimensional structure of H-Ras shows 6 beta sheets (shown in cyan) surrounded by 5 alpha-helices (in red). The binding site in this soluble protein is fairly exposed, in the outer surface above helix 2. This figure as well as the following representation of protein tertiary or quaternary structure as well as the binding site depiction (as below) have been prepared using the Accelrys DS ViewerPro software. Figure 5.6. The residues involved in the binding mechanism of Ras and residues in close proximity to the binding site are shown in the right image. GTP is shown in ball and stick. The Magnesium ion which is considered to be part of the binding site of Ras is represented with green ball. On the left is the depiction the binding sites surface coloured based on the Van der Waals electrostatics. Blue signifies electropositivity and is prominent on the right side of the site due to the two lysines (K147, K117). Red signifies electronegativity. The left side of the binding site shows electronegative potential due to Glutamic acid 31 and Aspartic acid 33 which form hydrogen bonds with the phosphate groups from ATP. For reasons of clarity the label of some amino acids has been omitted. 3.2.2.6.2 Target 2: Cyclooxygenase (COX-2) Biological Significance The formation of new blood vessels by angiogenesis to provide adequate blood supply is a key requirement for the growth of many tumours. It has been well established that Cyclooxygenase (also known as prostaglandin synthetase) inhibitors act as potent anti-angiogenic agents [38] therefore cutting down the essential blood supply to the cancer cells. Humans produce two different prostaglandin cyclooxygenases (namely COX-1 and COX-2) for different purposes. COX-1 is built in many different cells to create prostaglandins used for basic messages throughout the body. The second variant is built only in specialized cells and is used for signaling pain and inflammation. Unfortunately, the most of the current COX-2 inhibitors (with aspirin being the most famous of all as the most sold drug in history) attack both. Since COX-1 is targeted this can lead to unpleasant complications, such as gastrointestinal ulceration (stomach bleeding) and l toxicity. Fortunately, specific compounds that block just COX-2, leaving COX-1 to perform its essential jobs, are now becoming available these new drugs are selective pain-killers and fever reducers, without the unpleasant side-effects. However, as mentioned earlier, COX-2 inhibitors and anti-inflammatory drugs (NSAIDs) reduce the risk of colon cancer among others and that the inhibition of colon carcinogenesis by NSAIDs is mediated through the modulation of prostaglandin production [39] Colorectal cancer, one of the leading causes of cancer deaths in both men and women in Western countries, accounts for about 57,000 deaths annually in the United States alone [40] and is therefore a major public health problem. Several epidemiological studies have shown a significant inverse association between the intake of aspirin and the risk of colorectal cancer in the general population [41,42]. COX-2 overexpression has been reported not only for colorectal cancer though [43,44] but also in head and neck squamous carcinoma [45], oesophageal cancer [46, 47], skin cancer [48], gastric cancer [49] Protein structure and Binding site definition Each monomer of this dimeric structure contains 583 amino acids. An examination of the tertiary structure of the protein (see figure 8 below) reveals that COX-2 contains almost 40% helical residues and virtually no organized beta-sheets. Comparisons between COX-1 and COX-2 show that the two isoforms are 63% identical and 77% similar at the amino acid level [50,51]. The residues involved in heme binding and catalysis are highly conserved. The majority of differences occur in the mouth of the channel leading to the binding site, but seem to have little effect on the selectivity of NSAIDs. Many NSAIDs are competitive inhibitors, while some cause a time-dependent inhibition. The possibility of many different binding sites within the channel suggests that many sub-sites may exist for drug binding [48,51]. Identifying the sub-site differences between the two isoforms will be essential in creating NSAIDs that are COX-2 specific. There are three discrete folding units found in this molecule. The first (residues 33-72) is a compact domain held together by three intra-domain disulfide bonds. This unit is covalently linked to the rest of the protein by a disulfide bridge between cysteins 37 and 159. This domain is quite similar to that of the epidermal growth factor (EGF) thought to be responsible for initiating or maintaining protein-protein interactions. The intra-molecular disulfide bonds are conserved between COX-1 and COX-2 and occur between C36-C47, C41-C57, and C59-C69 [45-46;50]. The second folding unit (residues 73-116) contains four right handed helices that are believed to form the motif for insertion into the lipid bilayer [52]. The third and largest domain of the protein is the catalytic domain consisting of 466 residues (117-583). This folding unit contains the active sites for the cyclooxygenase and peroxidase activities. The catalytic domain contains a heme at the apex of the cyclooxygenase active site [52]. The catalytic domain is composed of two lobes. The larger lobe is built around helices H5 and H6 which grip H2. Packed around this core are H3, H10, H18, and H19. The smaller lobe is a bundle is formed by helices H1, H8, H12 and H14-H16 [50]. The binding site itself lies at the end of long hydrophobic narrow channel found on the outer surface of the membrane binding motif in the middle of three helices at the centre of each monomer. The walls of the tunnel are defined by four amphipathic alpha helices, formed by residues 106-123, 325-353,379-384, and 520-535 which are bound to the lipid membrane and give the channel dimensions of approximately 8 x 25 angstroms. Hydrophobic groups on specific sides of alpha helices allow connection of the membrane binding motif which leads to the hydrophobic channel and the cyclooxygenase active site. This allows molecules like fatty acids to directly reach the site [53]. Near the entrance of the channel is a valine at position 523. A COX-2 selective inhibitor called S58 (Figure 9) blocks this channel. This inhibitor prevents the binding of arachidonate, a fatty acid released from smooth ER membranes by phospholipase A2. Aspirin acetylates Ser 530 in COX-2. This is a very important catalytic residue [53]. This covalent modification occurs just below Tyr 385 blocking access of arachidonic acid into the upper portion of the channel [51]. Tyr 385 is also a catalytically important residue. Arachadonic acid is assumed to enter the active site in a bent conformation with C-13 in the vicinity of Tyr 385. Arg 120 which is the only polar residue in the channel besides Glu 524, is the ligand for the carboxyl group of the substrate. Regular NSAIDs inhibit both isoforms by binding to polar arginine at position 120 through hydrogen bonding and blocking the COX enzyme channel about halfway down the site. Heme-dependent peroxidase activity is implicated in the formation of a proposed Tyr-385 radical, which is required for cyclooxygenase activity and is therefore included in our definition of the binding site [45-46;50-51]. The other residues that interact with S58 have also been included in the binding site definition for this target and this include (His 90 and Phe 518 that form hydrogen bonds with the inhibitor and Gln 192, Val 349, Leu 352, SER 353, Leu 359 and al 523 that form van der Waals bonds with S58. The back of the channel binds a heme B cofactor that functions enzymatically to convert arachidonate into prostaglandin. A tyrosine residue, which is not present in the active sites of haemoglobin or nitrophorin, is critical for this enzymatic active site from inside the bilayer, this heme binding site is not included in the definition of this binding site as the antagonists would not bind there but lower down the channel. Figure 5.7. The three dimensional structure of Cyclooxygenase 2. This is a dimeric structure consisting mostly of alpha helices, loops and turns. The left monomer is coloured by secondary structure. The heme group present just above the binding site is coloured yellow. One can observe in the same region of the right monomer the channel where aspirin (or other inhibitors) would pass and acetylate Serine 530. Figure 5.8. Arginine 120 and Glutamate 524 are the only polar residues in this highly hydrophobic active site. The arginine tends to form hydrogen bonds with the carboxyl groups of the ligands but this doesn’t happen in the case of the inhibitor bound here (S58). A number of neutral and non-polar residues such as ALA 527, PHE 518 and partially TRP 387, offer hydrophobic interactions with the ligand (e.g. aspirin) to ensure a high affinity with the antagonist. Tyrosine 385 is also very important residue as it can block the way for the ligand traveling through the COX-2 channel. 3.2.2.6.3 Targets 3 and 4: Vascular Endothelial Growth Factor (VEGF) and Vascular Endothelial Growth factor Receptor 1 (VEGFR1) Biological significance VEGF also referred to as vascular permeability factor, is a dimeric glycoprotein that plays an important role in tumour angiogenesis a process necessary for tumour growth. VEGF is overexpressed in many human cancers, including prostate cancer [55,56], bladder cancer [57] neuroblastoma [58-60] and colorectal cancer [61]. VEGF levels were generally higher in the sera of the tumour patients compared to the sera of healthy control subjects [62] Overexpression of VEGFs is associated with angiogenesis of not only primary but metastatic cancers as well [63]. Human VEGF-A has at least six subtypes due to the alternative splicing of a single gene, VEGF121, VEGF145, VEGF165, VEGF183 , VEGF189, and VEGF206 (64, 65). Among them, VEGF165 has the most potent biological activity and is the most abundant subtype in vivo with a few exceptions such as in placenta and is the subtype examined for this project. VEGF-A binds to and activates two tyrosine kinase receptors. VEGFR is a family of VEGF receptors which also has multiple members. The family includes VEGFR-1 (Flt-1), VEGFR-2 (Flk-1/KDR), VEGFR-3 (Flt-4) [64,66]. In addition, VEGF165 also binds another membrane protein, neuropilin-1 (NRP-1) [67]. All of the vascular endothelial growth factor receptors belong to the group of receptor tyrosine kinases (RTKs) which typically consist of an extracellular domain that contains seven immunoglobulin-like regions, a transmembrane region, and an intracellular domain that contains a tyrosine kinase region [64]. VEGF is responsible for the activation of many diverse functions within the cell. Activation of the RTKs in the VEGFR family leads to promotion of endothelial cell mitogenesis, increased endothelial cell survival, chemotaxis, increased expression of proteolytic enzymes involved in stromal degradation, increased vascular permeability, inhibition of maturation of antigen-presenting dendritic cells, and vasodilatation[69]. Different RTKs in this family are linked to one or more of the preceding cellular functions. VEGFR-1 is thought to be important in migration [70,71]. The production of VEGF by tumors is known occur in response to various upstream factors, including hypoxia, elevated concentrations of bFGF, epidermal growth factor (EGF), insulin-like growth (IGF) and hydrogen peroxide (H2O2). Several lines of evidence show that VEGF is one of the most important factors in tumour cell survival and neovascularization [72]. For example, deletion of VEGF or its receptor in mice results in the loss of functional blood vessels and early embryonic lethality [72]. Furthermore, blocking VEGF or VEGF receptor functions can induce regression of tumour vasculature in vivo [72,73] as well as other problems associated with excessive angiogenesis such as rheumatoid arthritis, diabetic retinopathy, and age-related macular degeneration [74]. Therefore, angiogenesis has become a popular target for drug development, and several antiangiogenic compounds based on very different mechanisms of action are being developed and have been reviewed by V. Brower [75]. Protein structure and Binding site definition The crystal structure for this very important target was solved by C. Wiesmann and A.M. De Vos at a relatively high resolution (1.70Å) using X-ray diffraction. In this coordinate file the VEGF homodimer hormone is binding to the kinase domain receptor and the fms-like tyrosine kinase receptor (Flt-1) which is the VEGFR1 precursor (figure 9 below). The extracellular portions of the flt-1 consist of seven immunoglobulin domains only one of which (domain 2) is shown below. VEGF binds to and activates two tyrosine kinase receptors, kinase insert domain receptor (KDR) and fms-like tyrosine kinase receptor 1 (76). Most of the functions of VEGF are found to be mediated by KDR (77-79), and fms-like tyrosine kinase receptor 1 functions mainly as a decoy receptor, suppressing VEGF activity by capturing VEGF and thereby making it unavailable to KDR (78, 80-82). Activation of the VEGF receptors requires dimerization through binding of one receptor molecule to each of the two receptor binding sites on the VEGF dimer, located at the opposite ends of the VEGF dimer at the interface between the monomers (76). The crystal structure of the complex between VEGF and the second domain of Flt-1 shows domain 2 in a predominantly hydrophobic interaction with the "poles" of the VEGF dimer [19]. Here we examine the binding domain of both the VEGF as well as that of its receptor. According to Wiesmann et. al who solved this complex structure there is a number of predominantly hydrophobic interactions that stabilize the binding of VEGF to VEGFR1 which are formed between the pairs of residues as identified by BIND (Biomolecular Interaction Network Database) shown in table 2 below. Table 5.2. The 17 pairs of interacting residues between the VEGF and VEGFR1 interfaces VEGFR1 residue number 1. Glu 141 2. Ile 142 VEGF residue number Met 44 Phe 43 VEGFR1 residue number 10. Lys 200 11. Lys 200 VEGF residue number Glu 103 Arg 131 3. Pro 143 4. Pro 143 5. Lys 171 6. Phe 172 7. Tyr 199 8. Tyr 199 9. Tyr 199 Phe 43 Met 44 Tyr 51 Gln 48 Cys 130 Arg 131 Pro 132 12. Gly 203 13. Leu 204 14. Leu 221 15. Arg 224 16. Arg 224 17. Arg 224 Tyr 47 Tyr 47 Phe 43 Tyr 47 Asp 89 Gly 92 Previous analysis of the VEGF165 mutants for VEGFR-1 binding demonstrated that the most important VEGFR-1-binding determinants are Phe 17, Tyr 21, Gln 22, and Leu 66 [86-87]. All of the above mentioned residues that form protein-protein interactions between the hormone and its receptor are included in the definition of their respective binding sites. These two sites differ to those of most of the other targets in the sense that these are not well defined ligand binding cavities but relatively flat surfaces with very high accessibility which may potentially make the research for target specific leads harder in this case. In order to tackle this specificity problem it may be more relevant to examine possible allosteric binding sites for either of the proteins or focus the vHTS in the region of VEGF dimerization as the monomers are not active. Figure 5.9. The 3-dimensional representation of the VEGF homodimer (light green and yellow monomers) interacting with the binding domain of the VEGF receptor (Fms like tyrosine kinase domain shown here in dark green and bronze on either side of the hormone homodimer). It is important to notice that it is vital for the hormones’ activity to be present in a homodimeric form; therefore it may be of potential interest in the future to investigate ways to block the dimer formation instead of targeting the VEGR-VEGFR interface. Figure 5.10. The secondary structure of each VEGF monomer comprises of 4 β strands and 2 small helical domains. The β sheets of the two monomers are parallel to each other and form hydrogen bonds and hydrophobic interactions which help keep the 2 units joined together. The hormone interacts with the flt-1 receptor with the left side of this protein where mostly the α helical domain (residues 16-25) and the adjacent loop (residues 59-66) interact with the binding domain of the tyrosine kinase domain of the VEGF receptor. Figure 5.11. The VEGF interface presents a number of charged residues, namely 2 aspartic acids (Asp 19 and Asp 63) and a Glutamic Acid (Glu 103) which are represented by the negative areas (red on the left figure) and only a positively charged arginine (Arg105) in the bottom and right side of the site. The three neutral residues along the axis of the site (Phe 17, Tyr 21 and Tyr 25) are of great importance as well as the form hydrophobic contacts with residues from the receptor. Mutational studies have shown the alteration of Phe 17, Tyr 21, Gln 22 or Leu 66 as shown in the right diagram have as a result the decreased or non existent affinity between the two proteins. Figure 5.12. The structure of the Fms like Tyrosine kinase domain is also mostly made of β strands (2 antiparallel sheets with 4 and 3 strands respectively). VEGF interacts with the right side of the protein as it is depicted above. Figure 5.13. The receptors binding site is mostly neutral with the exception of the three basic residues (Lys 171, Lys 200 and Arg 224) and the two acidic (Glu 150 and Asp 175). These charged residues are involved in a number of interactions with the hormone (e.g. salt bridge of Lys 200 with Glu 103 from the VEGF) but other non-charged residues have also got important roles (e.g. Phe 172 which binds to Gln 48 and Tyr 51) T3.2.2.6.4 Target 5: Superoxide Dismutase Biological Significance Superoxide dismutase (SOD) is a critical enzyme responsible for the elimination of superoxide radicals and is considered to be a key anti-oxidant in aerobic cells. Cellular consumption of oxygen is essential for oxidative phosphorylation during ATP generation in the mitochondria, yet this cellular metabolism also leads to the production of reactive oxygen species (ROS), including the superoxide radical (O2 • - ) and hydrogen peroxide (H2O2). It was found over 30 years ago that SOD catalyzes the destruction of the O2- free radical to molecular oxygen and hydrogen peroxide: 2O2- + 2H+ O2 + H2O2 It has been well established that this enzyme protects oxygenmetabolizing cells against harmful effects of superoxide free-radicals [85-90]. McCord (1974) found that SOD protects hyaluronate against depolymerization inflammatory effect [91]. The O2- ion, which has been considered important in aging, lipid peroxidation and the peroxidative hemolysis of red blood cells [92], is formed by the univalent reduction of O2 during various enzymatic reactions or by ionizing radiation. [93]. SOD has been indicated to be present in all oxygen-metabolizing cells [94]. Three superoxide dismutases are characterized by different metal content. A blue-green Cu(II)-Zn(II) enzyme comes from human and bovine erythrocytes, a wine-red Mn(III) protein is found in E. coli, and in chicken, and rat [95] liver mitochondria [96] and a yellow Fe(III) enzyme from E. coli [97]. Bovine erythrocyte SOD which is where the crystal structure for this protein target has derived has been extensively studied. It is identical to the enzyme from human erythrocytes and from beef heart [98-100] Accumulation of ROS results in cellular oxidative stress and, if not corrected, can lead to the damage of important biomolecules such as proteins, DNA and membrane lipids. Salvemini et al suggested that active oxygen may be increased in tumour cells. [101] Extended accumulation of high levels of free radicals in cells may cause irreversible cellular injury and ultimately result in cell death. Since SOD is the key enzyme in the first metabolic step of superoxide elimination, deficiency in SOD or inhibition of the enzyme activity may cause severe accumulation of O 2 • - in cancer cells and lead to their death. Thus, inhibition of SOD may provide a novel way to kill cancer cells. Due to dysfunction in the regulation of cell growth, cancer cells are active in energy metabolism, and thus produce high levels of O 2 • - and other ROS and are under constant oxidative stress. This may render the malignant cells more dependent on SOD to eliminate the toxic superoxide radicals and thus potentially more sensitive to SOD inhibitors. It is a plausible hypothesis that inhibition of SOD may preferentially kill malignant cells through a free radicalmediated mechanism. [102] Protein structure and Binding site definition The structure for this target was solved by Lah et al. using X-ray crystallography at a resolution of 1.80Å. Superoxide dismutase consists of two subunits of identical molecular weight joined by a disulfide bond. The molecular weight is 32,500. There are two Cu(II) and two Zn(II) atoms per molecule. Rotilio et al. [103-105] have reported on the roles of copper and zinc. Zinc has a structural, stabilizing role [106], while Cu2+ is directly involved in the catalytic activity [107]. Forman et al. [106,108] and Rigo et al. [109] indicate His 26 to be involved at the active site. The azide in the binding site is covalently bound to the Fe and appears to hydrogen bond with His 30 and form a van der Waals interaction with His 31. Unfortunately though although this would theoretically be an ideal target for this project because of it biological activity, pharmacologically it is not so suitable. The 6 histidines that surround the binding site together with the large tryptophan and tyrosine create a very tightly packed binding site with extremely difficult accessibility (the only possible opening to the binding site is shown in figure 15) which is only big enough for a hydroxide or other very small inorganic compound. The actual binding site space is extremely limiting as well therefore this target was deemed unsuitable for this project. Figure 5.14. The quaternary structure of superoxide dismutase consists of 2 identical polypeptide chains. α helices are the dominant secondary structure characteristic throughout this protein although a β sheet made of three strand is present as well. The proteins small binding site is between the middle one of those strands and a small loop separating 2 α-helical domains. Figure 5.15. The superoxide binding site is a non polar site (with the exception of Asp 156) and has a very difficult accessibility as shown from the left figure. This is due to the 6 histidines, the tryptophan and the tyrosine that are surrounding it (right). 3.2.2.6.5 Target 6: Insulin Tyrosine Kinase (Insulin Growth Factor Receptor, IGFR1) Biological Significance The IGF system is complex and includes four different components. The first is the two growth factors and includes IGF-I and IGF-II, second is the two receptors and includes IGFR1 and IGFR2, the third is the six high-affinity binding proteins IGFBP1-6, and last is the IGFBP proteases. All these parts act in concert to regulate several cellular pathways in a wide variety of tissues throughout the body [110,111]. IGFR1 and insulin Receptors (IRs) are more frequently expressed on cancer in comparison to normal cells. These receptors form heterodimers after binding to IGF-1. This may lead to enhanced metastasis as IGFR1 activation leads to interaction with integrins. IGFR1 is more frequently expressed on tumors that are estrogen receptor (ER) positive[113]. Frigitta et al., observed that the insulin receptor content, as determined by a specific radioimmunoassay, was four fold increased in human breast cancer tissue when compared to normal breast tissues. IGFBPs also play a role in breast cancer. IGFBP-4 and -5 may prevent apoptosis in cancer cells, while IGFBP-3 inhibits cell growth both directly and by binding IGF-1, making it less readily available to bind the IGFR1. IGFR2 has no role in cancer cells and therefore was not chosen as a target for this screening project [114]. A great number of studies have shown that IGFRs are expressed in lung tumor tissues [115-117]. IGF-II overexpression has also been shown to be sufficient to induce lung tumourigenesis in an in vivo mouse model. While IGF-II levels are associated with a poor prognosis in pulmonary adenocarcinoma, prospective studies have not been able to demonstrate a link between IGF-I or IGF-II levels, and lung cancer risk [117]. However, high tumor IGF-II immunoreactivity has been correlated with decreased survival in patients with lung adenocarcinoma, suggesting that tissue-specific levels of IGF-II may be more relevant than plasma levels. The IGF system has been implicated in the pathogenesis as well as metastasis of malignancies and makes an ideal candidate for the research purposes of this project. Protein Structure and Binding Site Definition The crystal structure for this target was solved by means of X-ray diffraction by S.R. Hubbard at a resolution of 1.90Å and contains 192 residues. Two of those residues are mutated in comparison to the wild insulin receptor. The mutations are C981S and Y984F. The structure of IR has been solved both with and without the tyrosine kinase domain being phosphorylated (need that ref). The tyrosine kinase domain consists of an N-lobe (5 stranded beta sheet and 1 helix) and a C-lobe (8 helices and 4 beta strands). The activation of the IGFR1 begins with the binding of the ligand. This induces autophosphorylation and subsequent association with several other proteins leading to downstream signalling. Ligand binding causes a conformational change to the protein which is transmitted to the inside of the cell. This leads to ATP binding and phosphorylation of 3 tyrosines in the outside of the protein (actually present as PTP- phosphotyrosins in this instance of the protein), 2 tyrosines near the C-terminal and phosphorylation of soluble substrates such as insulin receptor substrate 1 and 2 (IRS-1, IRS2). The protein is highly inactive when no ligand is bound. In the inactive form the A loop sits in the active site with Tyr 1162 hydrogen bonding to Asp 1132. The active site is empty of ATP because the ATP binding site is occupied by the A loop and the N-lobe has a 30 degree upward tilt. This means that Tyr 1162 does not get phosphorylated. This is a very stable inhibitory conformation, which is necessary because the receptor exists as a dimer. Upon ligand activation the N-lobe moves 21 degrees down towards the C-lobe and Tyr 1158, Tyr 1162 and Tyr 1163 get phosphorylated and move the A-loop 30 Å out of the active site making it able to now receive ATP. The ligand co-crystallised with this protein is ANP. All the residues that interact with the ligand as well as those in their close vicinity have been included in the definition of the binding site for this target. More specifically, ANP forms hydrogen bonds with Gly 1005, Ser 1006, Val 1010, Lys 1030, Glu 1077, Met 1079 and ASP 1150. A number of van Der Waals interaction also occur between the ligand and the protein and this come from Leu 1002, Ala 1028, Asp 1083 and Asn 1137. Furthermore, ANP forms ionic bonds with the two magnesium ions (MG 301, MG 302) which form part of the binding site. Figure 5.16. The tertiary structure of the insulin receptor. On the left, the Nlobe consists of 5 beta sheet strands and an alpha helix close to the vicinity of the binding site. The C-lobe is on the right half of the molecule as depicted above and contains 8 alpha helices and 4 beta strands Figure 5.17. The Insulin Receptor binding site solid surface electrostatic display (left) reveals that the bottom half of the site is relatively non-polar (with the exception of Lys 1030 which forms a hydrogen bond with the ligand). The relatively spacious site contains other charged residues (e.g. Asp 1083, Asp 1150) which are involved in the electrostatic interactions with the ligand (ANP). The two magnesium ions present in the site are involved in ionic interactions with both the protein and the ligand. 3.2.2.6.6 Target 7: C-ABL Tyrosine kinase Biological Significance The biological activity of the c-Abl (Abelson) protein is related to its tyrosine kinase and DNA-binding activities The c-Abl proto-oncogene is located on the human chromosome 9 [130] and the encoded protein is ubiquitously expressed but is primarily localised in the cell nucleus [131] where unlike other tyrosine kinases, it binds to DNA and plays an important role in cell regulation and the cytoplasm. This unique function of c-Abl is regulated during cell cycle progression. c-Abl is activated by oxidative stress but its precise function in cell response to this stress is elusive [120] although there is nowadays growing evidence that this protein is very important in the cell cycle response to DNA damage[121]. The ubiquitously expressed c-Abl tyrosine kinase is activated in the apoptotic response of cells to DNA damage. The mechanisms by which c-Abl signals the induction of apoptosis are not understood [128]. It interacts preferentially with sequences containing an AAC motif and appears to exhibit a considerably higher affinity for bent or bendable DNA, as is the case with high mobility group (HMG) proteins. [129] Overexpression of c-Abl causes cell growth arrest in G1 phase [132,133]. It has been found that genotoxic drugs such as the antimetabolite ara-C and the DNA alkylating agent methylmethane sulphonate (MMS) activate c-Abl kinase activity leading to a complete G1 arrest in MCF-7 carcinoma cells [134]. This protein target is mostly related with Chronic Myelogenous Leukemia (CML), a myeloproliferative disorder. In this disease, the tyrosine kinase is improperly activated by the accidental fusion of the bcr gene with the gene encoding the intracellular non-receptor tyrosine kinase. This translocation leads to a shortened chromosome 22, called the Philadelphia chromosome. Overall, c-Abl was expressed in 71% of serous carcinomas. cAbl was expressed more frequently in the low-grade serous carcinomas (81%) compared with the high-grade serous carcinomas (65%) [135-136]. It is therefore clear that c-Abl tyrosine kinase makes an excellent target for this vHTS project as its inhibition could result in the cell cycle arrest of cancerous cells in a great proportion of malignancies. Protein Structure and Binding Site Definition The X-ray structure of this kinase target was solved by Nagar et al [22]. The coordinate file contains a homodimer of the proteins’ unphosphorylated kinase domain where each monomer is made of 293 amino acid residues. Nagar et al., have crystallized the protein with two different inhibitors namely STI-571 (also known as imatinib and Gleevec) and D173955. For this project we examined the structure crystallized with STI as this was solved at a higher resolution (2.10Å) in comparison to the other one with D173955 (2.60Å). Both ligands were bound in the canonical binding site of the ATP [22] Wild-type c-Abl is a relatively large protein (1150 residues). It has two distinct N- and C- termini domains. The N-terminal half (approximately 530 residues) of c-Abl has a high sequence identity to the Src family of tyrosine kinases (42% excluding the NH2-terminal unique domain) and shares a similar domain organisation, containing two modular peptide binding units (the SH2 and SH3 domains) followed by a tyrosine kinase domain. However, c-Abl is differs from the Src kinases as it is missing a crucial tyrosine residue that follows the kinase domain of c-Src. The carboxyl terminal half of c-Abl contains DNA and actin-binding domains mixed together with phosphorylation sites and other short recognition motifs, including proline-rich segments and nuclear localization signals [124] Under normal conditions, c-Abl exists in a regulated state with very low kinase activity [125], similarly to the IGFR1 described earlier. As cells enter mitosis, hyperphosphorylation of Ser/Thr residues inactivates DNA binding [122-123]. The DNA binding properties of c-Abl are not well understood. Initially, it was shown that the protein binds to a palindromic element, EP, of the hepatitis B virus enhancer [126,127]. A more recent study proposed that c-Abl binds essentially to A/T-rich DNA sequences via the cooperative action of three high mobility group 1-like boxes (HLB) [22] however in this study we are not researching the DNA binding site but the ATP site. The catalytic domains of eukaryotic Ser/Thr and tyrosine kinases are highly conserved both in sequence and structure. The kinase domain has a bilobal structure again similar to the IGFR1. The N-lobe (residues 225–350) contains a β-sheet and one conserved α-helix as shown in figure 17 below. The C-lobe (residues 354–498) is largely helical. The ATP binding site for this protein lies at the interface between these two lobes and is formed by a number of highly conserved residues. Small molecule inhibitors of protein kinases that have been discovered to date almost invariably bind to the kinase domain at this interfacial cleft between the two lobes, displacing ATP [22, 125]. Nagar et al in their study investigated the effect of several mutations in the binding affinity (in terms of IC50 values). We included all of the residues the mutation of which had a significant effect in the binding with STI. In addition, as per all targets, the residues that form direct interactions were also included in the defined vicinity of the binding site. These residues were Ala 269, Glu 286, Val 299, Thr 315, Met 318, Asp 381 that form hydrogen bonds with this ligand would also potentially bond with other antagonists, Leu 282, Lys 271, Met 290, Ile 313, Ile 360, Phe 382 that form van der Waals interactions and finally Tyr 253 and two phenylalanine residues (Phe 317, Phe 382) form two planes hydrophobic interactions with the ligand. Figure 5.18. The quaternary structure of c-Abl. The protein crystallized as a homodimer. The left monomer has been coloured according to secondary structure and the right monomer is coloured blue. The secondary structure differs between the C-lobe (the bottom half of the molecule as displayed above) which consists mostly of α-helices and the N a β-pleated sheet made of 5 strands is also present. The site where STI binds (displayed as ball and stick). Figure 5.19. The binding site of the c-Abl tyrosine kinase displayed with a van der Waals surface on the left and with the key amino acids in a stick display on the right. On the left diagram it is easy to notice that there are three distinct regions of polarity in this binding site. The left portion of the site is mostly electropositive due to an arginine and a glutamine, in the electronegative middle an aspartic acid and a glutamic acid come close to each other after the ligand is bound whereas the right portion of the site is largely non-polar. For reasons of clarity, a number of residues, certain side-chains and the labels of some aminoacids are missing from the right diagram. 3.2.2.6.7 Target 8: Fibroblast Growth factor Receptor 1 (FGFR1) Biological Significance The fibroblast growth factor (FGF) family of signalling molecules are mitogenic polypeptides which transduce signals through a class of cellsurface tyrosine kinase receptor called the Fibroblast Growth Factor Receptors (FGFRs) which contribute to normal developmental and physiological processes. It is therefore of no surprise that any changes in this powerful growth stimulatory pathway have been implicated in the generation of a variety of pathological conditions [136]. This signalling pathway is associated with and functionally important for the growth of some human tumours. In mammals, the FGFs constitute a large family of more than 20 structurally homologous ligands. The FGF receptors are encoded by four genes (FGFR1 to FGFR4) that give rise to seven prototype receptors because of alternative splicing. Upon binding of the ligand, FGF receptors homodimerize or heterodimerize to activate various intracellular signaling pathways. FGFs have been identified as oncogenes in murine mammary cancer and FGF receptor genes have been reported to be amplified in some human breast cancers [137,138] Pancreatic cancers overexpress basic FGF (bFGF) and the type I FGF receptor (FGFR-1), and overexpression of bFGF has been correlated with decreased patient survival. [139,140] The other type of cancer where the FGFR1 is highly documented to be over expressed in a substantial fraction of cancer patients is prostate cancer. [104,141,110] Fibroblast growth factors (FGFs) play an important role in the growth and maintenance of the normal prostate. There is increasing evidence from both animal models and analysis of human prostate cancer cell lines that alterations of FGFs and/or FGF receptors (FGFRs) may play an important role in prostate cancer progression. In addition, we have observed overexpression of both FGFR-1 and FGFR-2 in the prostate cancer epithelial cells in a subset of prostate cancers and that such overexpression is correlated with poor differentiation. [142]. Finally, FGFR1 is also found to be overexpressed in Glioblastoma Multiforme Tumors [143,144]. The FGFR-1 was selected as a target for this project because of the fact that it is found both to be overexpressed in a number of common lethal human carcinomas as well as associated with the extensive spread of these cancerous growths, however inhibitors of this target would also be of clinical importance as mutated FGFRs are known to cause skeletal disorders. Protein Structure and Binding site Definition The structure for this target was solved by means of X-ray crystallography by M. Mohammadi and S.R. Hubbard (who also crystallized the IGFR1 receptor discussed above) at 2.40Å and contains 310 residues of the tyrosine kinase domain of the Fibroblast Growth Factor Receptor (FGFR1K) in complex with an inhibitor of the protein (SU4984) which is based on an oxindole core which occupies the ATP binding site. Very similarly to the IGF receptor 1 and the c-Abl tyrosine kinase described earlier, the FGFR1 receptor also consists of an N-lobe and a C-lobe and has an A-loop in close proximity to the binding site. But the A-loop in this case is not as hindering as in the case of the IGFR1 and ATP access is not restricted. Furthermore, these two lobes are not tilted as far apart as is the case for the IGFR1. FGFR therefore exists in a less stabilised inhibitory conformation. This is because a high level inhibition is not required as the inactive form is a monomer. However as in the case of the IGFR1, the C- terminus lobe of this tyrosine kinase domain of the FGFR1 is mostly α helical whereas the N- terminus domain has a large β sheet made of 5 antiparallel strands. Oxindole based inhibitors (indolinones) bind to the same region of FGFR as ATP and therefore inhibit the receptor. The oxindole of the inhibitors occupies the same region of the site as the ATP adenine, although the orientations of the bicyclic ring systems differ by nearly 180°. The chemical groups attached to C-3 of the oxindole emerge from the cleft at approximately right angles to the direction taken by the rest of the ATP molecule. Neither inhibitor binds near the putative substrate peptide binding site in the COOHterminal lobe of the kinase, indicating that these inhibitors do not compete with substrate peptide. The oxindole forms two hydrogen bonds to the receptors backbone in the kinase region FGFR1. These are between the N-1 of the oxindole and the carbonyl oxygen of Glu 562, and between O-2 of the oxindole and the amide nitrogen of Ala 564. These two particular residues are present in the hinge region, the segment between beta strand 5 and α helix D (residues 563 through 568) that connects the two lobes of FGFR1K. These same two backbone groups of FGFR1K make hydrogen bonds to N-1 and N-6 of the ATP adenine. The cavity in which the oxindole (or adenine) binds is surrounded with a number of hydrophobic residues including Val 492, Ala 512, Ile 545, Val 561, Ala 564, and Leu 630. In addition, Leu 484 and Tyr 563 provide a hydrophobic environment for the ring proximal to the oxindole—a phenyl in SU4984 and a pyrrole in SU5402 (a similar FGFR1 inhibitor of the oxindole family) which shows that these 2 residues are crucial for the binding of ligands and were therefore included in the binding site definition of this target (figure 20 below)[145,147]. The phenyl ring of SU4984 makes an oxygen-aromatic contact with the carbonyl oxygen of Ala 564. The piperazine ring of SU4984 is in van der Waals contact with Gly 567, a highly conserved residue in protein kinases. According to Hubbard et al, the terminal formyl group of the inhibitor SU4984 has poor associated electron density which indicates that this group is disordered. Indeed, a compound lacking the formyl group is as potent an inhibitor as SU4984 (146). In the GFR1K - SU5402 structure, N-19 of the pyrrole ring makes a hydrogen bond with the O-2 of the oxindole. The methyl group of the pyrrole ring is in van der Waals contact with Gly 567, and the carboxyethyl group attached to C-39 of the pyrrole ring is hydrogen-bonded to the side chain of Asn 568, the last residue in the hinge region. Asn 568 is likely to be involved in ATP binding, because the corresponding residue in the insulin receptor tyrosine kinase (Asp 1083 see figure x above) makes a hydrogen bond with one of the ribose hydroxyl groups [145,147]. All of the above mentioned residues which according to the literature published by Mohammadi et al are involved in the binding of oxindole based inhibitors as well as the following residues which were also determined to be involved in intermolecular interactions between the ligands and the receptor have been included in the defined binding cavity for this site. The interactions observed which were not mentioned above include the hydrogen bonds between the O-2 and the N1 of the oxindole ring with Glu 562 and Tyr 563 respectively as well as the hydrogen bond of Ser 565 with N4. It is interesting to notice that Tyr 563 only forms a hydrogen bond with the ligand in the A subunit of the homodimer, in the B subunit it is tilted in a way making a 2 plane interaction with the phenol ring. Figure 5.20. The quaternary structure of the FGFR1 above shows how the receptor monomers interact to form the homodimer. The A subunit is coloured according to secondary structure and the B subunit is coloured blue. The binding in each monomer is located in close proximity to the loop which is located between the N and the C termini and joins the two lobes together similar to other kinases examined earlier. Figure 5.21. The FGFR1K binding site is lined with a number of hydrophobic residues namely Val 492, Ala 512, Ile 545, Val 561, Ala 564, and Leu 630 which form hydrophobic interactions with the four rings present in most of the receptors specific inhibitors like SU4984 represented here in balls and stick. There is a lack of strong electrostatic interactions as the only charged residues in this binding site are Lys 482 and Glu 568 which are approximately 4.5Å away from the O-3 of the piperazine ring. Some hydrogen bonds (eg Ser 565 with SU4984) also help increase the affinity of the binding. 3.2.2.6.8 Target 9: Cyclin dependent Kinase 2 (CDK-2) Biological Significance Cyclin-dependent kinases (CDKs) are enzymes controlling the eukaryotic cell cycle. It is common knowledge that ATP-dependent phosphorylation of serine or threonine residues in proteins alter the function of the protein. Changes in the activity of protein kinases can therefore regulate the cell cycle. CKDs, as their name suggests are activated by cyclins. Their activity is regulated by complex mechanism that includes binding to positive regulatory subunit and phosphorylation at positive and/or negative regulatory sites [148]. CDK-2 requires binding to cyclin A or cyclin E for activation. Specifically, CDK2 in association with cyclin E promotes the transition between G1 and S phase, and in association with cyclin A promotes progression through and exit from S phase. For full activity, CDK2 requires phosphorylation of a threonine residue in the activation segment of the kinase (Thr160) in addition to association with a cyclin molecule. The structural basis of CDK2 activation mechanisms [149], substrate specificity [150,151] and small molecule inhibitor recognition [152,153] are well understood. Another family of regulatory proteins, the cycle-dependent kinase inhibitors (CKIs) have the exact opposite effect of CDKs. Interactions between these three classes of proteins the regulates the checkpoints of each cell cycle as shown in figure 5 below. Cyclin CKI CDK Hypophosphorylated Serine substrate Hyperphosphorylated Serine substrate Phosphatase Figure 5.22. The Cyclin-Dependent protein kinase system. CDKs are activated by cyclins and inhibited by CKIs. When activated they phosphorylate other proteins (e.g. Retinoblastoma protein) which will then decide the fate of the cell cycle. As cyclins and CDKs play key roles in cell cycle regulation they make important cancer research targets as any deregulation to the cell cycle for example by increased expression of activating cyclins (e.g. cyclin D), decreases expression of inhibitory peptides (e.g. p16 and the Ras protein p21), or substrate loss (e.g. Rb gene deletion) can lead to uncontrolled cell growth and hence to cancer [154]. The inhibition of cyclin-dependent kinases has therefore been a topic of major interest in current anti-cancer drug research for a substantial amount of time. Different classes of chemical inhibitors of these enzymes have been identified during the past decade and the structural basis of inhibition has been elucidated by X-ray crystallography studies of CDK-2. Protein Structure and Binding Site Definition The crystal structure for CDK-2 was solved by Endicott et al., at 2.0Å by means of X-ray crystallography and the coordinate file contains 298 amino acid residues complexed with the antagonist staurosporine. CDK-2 features a typical kinase fold as described earlier for the c-Abl tyrosine kinase, i.e. the Nand C- termini are linked by the hinge region. The N-terminal lobe consists of 5 antiparallel β-strands and one α helix called the C helix (see figure x below). There exists a glycine rich loop which is located in the loop between strands β1 and β2 and is flexible due to the small size of the glycines. The C- terminal lobe is mainly α helical and is connected to the N-terminal domain via the hinge region. The C- terminal lobe also includes the activation segment (Tloop), the stretch of chain that runs between the conserved DFG and APE motifs in protein kinases and which carries the phosphorylated threonine residue, pThr160 [155]. CDK-2 resumes full activity by phosphorylation of the Thr160 residue in the activation segment [160]. The protein’s activity is natively inhibited mainly with two different ways. Either by dephosphorylation of the phosphothreonine or as this projects investigates with interactions with various natural protein inhibitors [161] However it has been established that CDK2 can be negatively regulated by phosphorylation at Tyr15 and (theoretically) at Thr14 [162]. The ATP substrate-binding site which in this instance is occupied by staurosporine is located between the two lobes. The adenine moiety of ATP makes two crucial hydrogen bonds to main chain atoms of the hinge region. The N1 atom of the adenine hydrogen bonds to the main chain nitrogen of Leu83 while the N6 group hydrogen bonds to the main chain oxygen of Glu 81 (see figure x below). The x-ray crystal structure of CDK2 has also been complexed with a 4-anilinoquinazoline was determined and is the first reported experimental determination of binding mode for that important class of protein kinase inhibitors. The inhibitor was found to bind in the ATP site, and the interaction between the quinazoline and CDK2 was mediated by a hydrogen bond linking quinazoline N1 to the backbone NH of Leu-83 (need to clear up that figure for L83). Staurosporine, a nonselective kinase inhibitor with nanomolar Ki for many protein kinases, has been shown to bind in almost identical modes to four different kinases [cyclin-dependent protein kinase 2 (CDK2) [156], cyclic AMP-dependent protein kinase (PKA) [157], C-terminal Src kinase (CSK) [158] and leukocyte specific kinase (LCK) [159]. As with all of the protein targets in this project, the residues that interact either with the natural ligand either via a hydrogen bond like Leu 83 and Glu 81 which were already mentioned (together with Ile 10, Phe 82 and Gln 131) as well as the ones that form hydrophobic interactions and van der Waals interactions (Gly 13, Ala 31, Lys 33, Asp 86, Leu 134 and Asp 145) including their important close neighbors are included in the area of the binding site as for example the ‘bulky’ Phe 80 which has been reported to shift staurosporine by 1.2 Å out of the potential hydroxy-group-binding pocket. Certain interactions between the ligands and the CDK2 binding sites (e.g. the interaction of the 7-hydroxy group on the UCN-01 inhibitor) have been suggested to be was described to be water-mediated in CDK2 due to the lack of hydrogen-bonding residues [163]. Other residues which have been previously reported to interact with different inhibitors were also present in our definition [155]. However, for the purposes of this project all the water molecules inside the binding site vicinity were removed in the protein preparation stages and therefore their possible biological effect there was not simulated. Figure 5.23. The three dimensional structure of CDK-2 is quite typical of kinases. There are two distinct domains, the N- terminus lobe on the left consists of a β sheet made of 5 antiparallel strands and an α helix called the C helix and the C-terminus which is mostly α helical. In between the two domains lies the binding site where there is a conserved loop which contains crucial residues for the catalytic machinery of this target. Figure 5.24. The binding site for this particular target is a well defined, mostly hydrophobic in its interior (apart from an Asp) pocket. The cavity is large enough to accommodate Staurosporine (shown as a ball and stick model) a large, non-specific kinase inhibitor which targets the ATP binding site. This site has a good accessibility for such a non-flexible ligand. Staurosporine is a large, rigid, planar ligand which interacts in a number of ways with its surround residues. 3.2.2.6.9 Target 10. RAF-1 Biological Significance It has nowadays become common knowledge that the deregulation of the Ras/Raf/MEK/ERK pathway (e.g. figure 6) plays a major role in cancer pathogenesis. Raf-1 is activated by a wide range of growth factors and hormones, and is a point of integration for numerous external signals. Hence, it is not surprising that the activation of Raf kinases is complex and still incompletely understood. Most of the stimuli that activate Raf-1 also activate Ras which was discussed earlier (target 1) [164-167]. FGF FGFR-1 Cell membrane P Grb2 SOS RASGTP RAF RASGDP GAP MEK ERK ERK Cytosolic Substrates Nucleus Transcription Figure 5.25. An example of RAF activation from RAF which in turns leads to activation of the mitogen activated ERK activating kinase (MEK), and extracellular signal regulated kinase (ERK). Upon activation, the ERKs phosphorylate cytoplasmic targets and they also translocate to the nucleus where they stimulate gene expression through the activation of transcription factors Raf-1 is a MAP kinase kinase kinase (MAP3K) which functions downstream of the Ras family of membrane associated GTPases to which it binds directly. Once activated Raf-1 can phosphorylate to activate the dual specificity protein kinases MEK1 and MEK2 which in turn phosphorylate to activate the serine/threonine specific protein kinases ERK1 and ERK2. This protein is ubiquitously expressed, whereas A-Raf is predominantly found in urogenital tissue and B-Raf shows the highest expression in neural tissue and testis [168]. All three Raf proteins also share common mechanisms of activation and downstream effectors. They are all activated by Ras and in turn activate the very complex mitogen-activated protein kinase/extracellularsignal-regulated kinase (MAPK/ERK) pathway by phosphorylating the MAPK/ERK kinase (MEK), which activates MAPK/ERK [164-165]. It has been recently shown that signaling events mediated by the basic Fibroblast Growth Factor (bFGF) in endothelial cells targets Raf-1 to the mitochondria, which protects these cells from apoptosis [168] This provides a mechanism that effectively explains why targeting the tumor neovasculature with a mutant Raf-1 gene exerts anti-angiogenic effects [169,170]. Recently, Alavi et al. demonstrated that bFGF and VEGF utilize the same target, i.e. Raf-1, kinase with distinct specificity [168] These studies suggest new possibilities for targeting the tumor neovasculature with small molecule drugs directed against Raf-1 that could promote the apoptosis of endothelial cells and cause regression of tumor vasculature. Endothelial cell death also plays a key role in myocardial infarction and heart failure. These studies also suggest opportunities for inducing therapeutic angiogenesis, in tissues where unwanted apoptosis could be prevented by promoting the translocation of activated Raf-1 kinase into the mitochondria. Several mechanisms have been proposed to explain how Raf can prevent apoptosis [171,172] It is commonly known that most cancer patients do not die from the primary tumour, but from the cancer’s metastases [173]. The ability to invade adjacent tissues and eventually spread into remote parts of the body epitomizes the malignancy of a cancer cell. About 50% of metastatic tumours feature activating Ras mutations. Webb et al in an ingenious study used g NIH 3T3 fibroblasts transfected with Ras mutants that selectively activate different downstream effectors, it was shown that all Ras mutants formed tumours in mice, but only a Ras mutant able to activate Raf supported metastasis. Cells re-isolated from the tumours and metastases exhibited higher levels of active ERK than did the cells originally injected, suggesting that a selection for higher ERK activity had occurred in vivo [173]. From all of the above it is obvious that RAF1 was chosen to be one of the targets for this research project both because of the fact that it is a downstream effector of Ras as well as because of its ability to prevent apoptosis and facilitate the metastasizing of tumours throughout the body. Protein Structure and Binding Site Definition The structure for this target was solved by X-ray diffraction by N. Nassar at 1.90Å and it contains 244 amino acid residues in total. Out of those, 167 residues actually belong to the Ras-related protein Rap1A and only 77 belong to the kinase domain of this proto-oncogene serine/threonine kinase target of this heterodimeric crystal. There are two binding sites associated with this target. These are that of the Rap1A which presents a very high similarity to that of Ras itself as well as the interface between Rap1A and the Ras-binding domain of raf-1. The three Raf proteins share a common structure consisting of an Nterminal regulatory domain and a C-terminal kinase domain. There are three conserved regions (CR1–CR3) of homology: in the regulatory domain, CR1 harbours a Ras-binding domain (RBD) and a cysteine-rich domain, and CR2 is a serine/threonine-rich domain; CR3 is sited in the kinase domain and is required for Raf activity. The removal of the regulatory domain generates an oncogenic kinase, which in the case of Raf-1 is often referred to as BXB [164,165]. The activity of Raf-1 can be modulated by both Ras-dependent and Ras-independent pathways. Arg89 of Raf-1 is a residue required for the association of Raf-1 and Ras. Mutation of this residue disrupts interaction with Ras and prevents Ras-mediated, but not protein kinase C-or tyrosine kinasemediated, enzymatic activation of Raf-1. Further analysis of this mutant demonstrates that kinase-defective Raf-1 proteins interfere with the propagation of proliferative and developmental signals by binding to Ras and blocking Ras function. These findings have also shown that phosphorylation events play a role in regulating Raf-1. Sites of in vivo phosphorylation that positively and negatively alter the biological and enzymatic activity of Raf-1 have been identified. Some of these phosphorylation sites are involved in mediating the interaction of Raf-1 with potential activators (Fyn and Src) and with other cellular proteins [174] The Raf-1 serine/threonine kinase is a key protein involved in the transmission of many growth and developmental signals. Autoinhibition mediated by the noncatalytic, N-terminal regulatory region of Raf-1 is an important mechanism regulating Raf-1 function. The inhibition of the regulatory region occurs, at least in part, through binding interactions involving the cysteine-rich domain. Events that disrupt this autoinhibition, such as mutation of the cysteine-rich domain or a mutation mimicking an activating phosphorylation event (Y340D), alleviate the repression of the regulatory region and increase Raf-1 activity. Based on the striking similarites between the autoregulation of the serine/threonine kinases protein kinase C, Byr2, and Raf-1, it is proposed that relief of autorepression and activation at the plasma membrane is an evolutionarily conserved mechanism of kinase regulation [175,176]. The first binding domain to be analysed here is that of the GTP site in the Rap1A (figure 27 below). The reason for investigating this binding site is primarily in order to cross-validate the returned hits from the Ras target as the ligands that bind to Rap1A should dock with a high affinity to Ras as well. All the residues that interact with the GTP analogue molecule in the Rap1A binding site have been included in the definition for this binding site. There is an extensive number of amino acids that form hydrogen bonds with this ligand and these include Gly 12, Gly 15, Lys 16, Ser 17, Ala 18, Glu 30, Tyr 32, Pro 34, Thr 35, Gly 60, Asn 116, Lys 117, Asp 119 and Ala 148. Gly 13 and Val 27 form weak van Der Waals bonds interactions and finally the aromatic ring of Phe 28 forms a two plane interaction with the adenine ring of ATP. The second site investigated for the purposes of this vHTS project was the Rap1A- Raf1 interface on the RBD of Raf1. Yeast two-hybrid and in vitro binding studies showed that Raf-1 residues 55-131 are sufficient for stable association with Ras [177-178]. Furthermore, structural studies conducted with both Ras and the Ras-related protein, Rap1A, indicate that Raf-1 residues 55-131 interact with residues 33-41 in the Ras effector region [25,179]. Finally, the critical role for Raf residues 55-131 in Ras-mediated activation of Raf-1 is established by the ability of a point mutation of Arg 89 to Leucine as the Raf (R89L) in the mutant the Ras-Raf-1 binding and Raf-1 kinase activation are disrupted [180] as this enormously important amino acid residue interacts with 4 different residues of Rap (3 hydrophobic interactions and 1 hydrogen bond). All of the residues in the Raf-1 interaction surface that interact with Ras or Rap according to literature have been included and are shown in table 3 below as identified with BIND. Table 5.3. The 23 pairs of residues that interact with each other during the Raf1 activation from Rap1A. Certain residues interact with more than one of the aminoacids of the other polypeptide chain. It is important to notice that the RBD of Raf1 has a greater interaction surface in comparison to the Ras analogue and therefore it was the surface chosen for screening for an inhibitor or Raf1 activation. Raf1 residue number 1. Thr 57 2. Arg 59 3. Asn 64 4. Lys 65 5. Gln 66 6. Arg 66 7. Gln 66 8. Arg 67 9. Arg 67 10. Arg 67 11. Thr 68 12. Thr 68 Rap1A residue number Ile 36 Glu 37 Arg 41 Arg 41 Arg 41 Tyr 40 Arg 41 Glu 37 Asp 38 Ser 39 Glu 37 Asp 38 Raf1 residue number 13. Val 69 14. Val 69 15. Asn 71 16. Lys 84 17. Val 88 18. Val 88 19. Arg 89 20. Arg 89 21. Arg 89 22. Arg 89 23. Gly 90 Rap1A residue number Ile 36 Gln 25 Ile 36 Asp 33 Gln 25 Tyr 40 Gln 25 Asp 38 Ser 39 Tyr 40 Gln 25 However it has been observed that mutations outside the Ras effector domain can also impair Ras-Raf-1 binding and Ras-mediated cell signaling [181-183] suggest that other Ras recognition elements may contribute to Raf1 kinase regulation. In particular Drugan et al [184] have identified our identification of a second Ras-binding site in the Raf-1 cysteine-rich domain (residues 139-184 which unfortunately Nessar et al did not crystallize and therefore could not be investigated. It is undoubtable that when a crystal structure for the Raf-1 domain is established, a significant amount of research will be spent in examining the alternative interaction sites between the two proteins as well as possible allosteric that would inactivate Raf-1. Figure 5.26. The larger domain on the left (167 amino acid residues) is that of the Ras-related protein Rap1A and on the right the smaller kinase domain of Raf-1 (77 amino acid residues) present in the RBD of the protein. The binding site of the Rap1A is largely accessible to the solvent as it is the bottom area of the protein (as depicted here) that is anchored to the cell membrane after farnesylation. Figure 5.27. The Rap1A binding site presents a high similarity to that of HRas discussed earlier. GTP enters the well defined cavity and engages strong interactions with several residues in the vicinity of the active site cavity. As RAP1A belongs to a family of RAS-related proteins, it is no surprise that these proteins share approximately 50% amino acid identity with the classical RAS proteins and have numerous structural features in common. The most striking difference between the RAP and RAS proteins resides in their 61st amino acid (Phenylalanine in Rap, Glutamine in Ras) however a number of other residues in the binding site differ as can be established by compairing the above figure with figure 6. Figure 5.28. An examination of the surface of the Ras Binding domain of Raf1 interface with Ras shows a highly electropositive surface due to the large number of basic residues (4 arginines and 1 lysine). On the opposite side, the the significantly smaller Rap1A protein-protein interaction surface (not shown) shows a great electronegativity due to three acidic residues (2 aspartates and one glutamate). 3.2.2.6.10 Target 11. Farnesyltransferase Biological Significance The significance of the Ras protein and its relation to cancer was described earlier on. Like every G-protein, it is essential for both normal as well as mutated Ras proteins to be bound to the cell membrane to ensure the signal transduction takes place. The anchorage of Ras proteins to the cell membrane is accomplished by a series of posttranslational modifications (altogether known as prenylation) modulated by this enzyme target, farnesyltransferase (FTase). The addition of the 15-carbon prenyl or farnesyl (shown in figure x below) moiety by farnesyl transferase is critical to the function of a number of other proteins as well [186,187]. FTase catalyzes the attachment of farnesyl, through a thioether linkage to a cysteine near the C-terminus of oncogenic Ras proteins. These transform animal cells to a malignant phenotype when farnesylated. This amino acid is a component of the so called "CaaX-box" located at the C-terminus of Ras, where "C" stands for cystein, "a" for an aliphatic amino acid and "X" is methionine, serine, alanine, or glutamine (depending on the Ras protein). Since FTase is responsible for the activity of oncogenic Ras proteins, it is an interesting target for the development of antitumour agents. It is now clear that farnesyl transferase inhibitors (FTIs) have activity independent of Ras, most likely due to effects on prenylated proteins downstream of Ras, which explains their activity in several malignancies, including breast cancer, where Ras mutations are rare. Therefore, although originally developed as inhibitors of RAS signal transduction pathways, it is now apparent that these drugs are better described as prenylation inhibitors. [186-188] Figure 5.29. The 15 carbon farnesyl (prenyl) moiety which farnesyl transferase adds to Ras and other oncogenic and not proteins regulating their binding to t. Several FTIs are in clinical development for the treatment of solid tumours. Pre-clinical evidence suggests that FTIs can inhibit breast cancers in vitro and in vivo, and a phase II trial of the orally administered FTI R115777 in patients with advanced breast cancer, produced encouraging results. [189]Apart from a number of lead structures, showing potent inhibitory activities, have been investigated, also a significant number of FTIs are currently undergoing clinical evaluation [190-195] which demonstrates in a striking way the potential of this strategy to treat malign illnesses. Protein Structure and Binding Site Definition The 3-dimenstional structure coordinates for this protein target were deposited by Long et al. who solved this protein by X-ray crystallography at 2.0Å. The protein consists of 814 residues (377 in the subunit and 437 in the β subunit). 11 residues of the K-Ras loop to be farnesylated has also been crystallized as well as the ligand which is an FPP analogue (PPT). Both units are highly α helical with no organised β strands present as it can be seen in figure x below. In the α subunits of both types of protein prenyltransferases, seven tetratricopeptide repeats are formed by pairs of helices (helices 2 to 15) that are stabilized by conserved intercalating residues. The tetratricopeptide repeats of the α subunit form a right-handed superhelix, which embraces the (α-α)6 barrel of the β subunit. The β subunits include most of the substrate and lipid-binding pockets [196] and their tight association with the respective α subunits is required for proper function [197]. The mode of binding in the active site of the FTase was investigated with the help of X-ray crystal structures of crystallized complexes from FTase, FPP and a CaaX mimetic [198-199]: A central zinc ion promotes the deprotonation of the cystein to its thiolate which in turn substitutes the pyrophosphate group of FPP generating a C-S linkage and thus connecting the farnesyl residue to the Ras protein. In the next steps the "aaX" part is cleaved off by a specific endoprotease and the free S-farnesylcystein at the C-terminus is finally methylated with the help of a methyltransferase enabling the Ras protein to bind to the cell membrane. [206] . Detailed mechanistic studies have been carried out to elucidate the catalytic mechanism of FTase. [196] Early work established the requirement for zinc in the catalysis for PFT [200,201]. The presence of these metals is essential for the catalytic machinery of this enzyme to work properly as it has been shown that Zn is directly coordinated to the cysteine thiolate (thiol) of the bound peptide substrate and to the thiol of the thioether-linked product and this supports a mechanism by which Zn enhances the nucleophilicity of the cysteine thiol of the protein substrate CaaX motif for the attack at the C1 carbon of FPP. Substrate binding in farnesyltransferase has been well characterized for both farnesyl and peptide substrates. The binding site is primarily located in the βsubunit however a significant number of amino acid residues from the αsubunit have also being involved in FPP and peptide binding. The isoprene tail of FPP binds deeply in the active site cavity at the centre of the β-subunit α-α barrel. Its diphosphate head binds to a positively charged cluster near the subunit interface. The positively charged side chains of residues Lys 164α, Tyr 166α, His 248β, Arg 291β, Lys 294β, as well as the Tyr 300β side-chain hydroxyl interact with the α- and β- phosphates of FPP and also form hydrogen bonds with the ligand (Figure x). The hydrophobic isoprene tail of FPP contacts many hydrophobic residues that line the funnel-shaped substrate binding pocket, including Trp 102β, Tyr 154β, Tyr 205β, Cys 254β, Tyr 251β, His 248β, Trp 303β, and Tyr 166α. The third isoprene unit packs against the aliphatic portion of Arg 202β side chain as well as the Ile side chain of the CVIM (the CaaX mentioned earlier) tetrapeptide moiety in the reactant ternary complexes.[202-204] Figure 5.30. Ribbon diagrams of rat protein farnesyltransferase, The α domain is coloured based on its secondary structure and the β-subunit which consists of 7 pairs of helices is coloured blue. The K-Ras loop which consists of 11 amino acids is displayed in yellow. The Zn ion and the protein ligand (FPP analogue) are represented in ball and stick. The binding site lies between the α and the β subunit and residues from both these domains interact with the ligand, shown in ball and stick in the middle of this figure. Figure 5.31. The binding site of farnesyltransferase is made of residues from both its α and its β subunits. An examination on the electrostatics of this binding site (left) shows that this is a highly electropositive site mostly due to the presence of the basic lysines, arginines and a histidine. These specific residues are shown on the right together with other that surround the binding site area and/or have been shown to interact with the FPP ligand or its analogue shown here. 3.2.2.6.11 Target 12: Protein Tyrosine phosphatase 1b (PTP1b) Biological Significance The phosphorylation and de-phosphorylation of tyrosine residues in proteins, catalysed by the antagonistic coordinated actions of protein tyrosine kinases and protein-tyrosine phosphatases (PTPs), is of vital importance to the control of basic physiological functions as cell proliferation, differentiation, survival, metabolism, and motility. Protein Tyrosine Phosphatase works antagonistically to all of the kinases already mentioned above as shown in the next diagram. PTK Tyrosine residue ATP Phosphotyrosine residue ADP P OH Protein A ADP ATP PTP P Phosphorylated Protein A Figure 5.32. Tyrosine phosphorylation is a reversible, dynamic process controlled by the activities of the protein tyrosine kinases (PTKs) and the competing actions of the protein tyrosine phosphatases (PTPs). Phosphorylating a protein enzyme usually results in a significant change in its shape, activity and kinetics. PTKs phosphorylate tyrosine residues on a substrate protein and PTPs remove these phosphates from substrate tyrosine. Since the phosphorylation status of a protein can modulate its function, PTKs and PTPs work together to regulate protein function in response to a variety of signals, including hormones, mitogens, and oncogenes. [205-207]. The phosphorylation of a target protein alters its function, including changes in enzymatic activity or its ability to associate with other proteins. In response to a stimulus, such as a growth factor or hormone, multiple phosphorylation and dephosphorylation reactions are coordinated in signal transduction cascades that culminate in the physiological response [208-209] An extensive amount body of information has been built up to describe the role of protein tyrosine kinases in the regulation of signal transduction. However, we are only now beginning to appreciate in mechanistic detail the role of some members of the PTP family in fine-tuning the signalling response to extracellular stimuli [205]. Analysis of the human genome sequence revealed the existence of 38 PTP genes in humans [210-211] These PTPs compose receptor-like proteins, which have the potential to regulate signalling directly through ligand-controlled protein dephosphorylation, as well as nontransmembrane, cytoplasmic enzymes. In addition, there are 60 dualspecificity phosphatases, which are members of the PTP family that recognize Ser/Thr and Tyr residues in proteins [208] making a total of approximately 100 phosphatases that belong to the protein tyrosine phosphatase (PTP) superfamily. The substrates for PTPs range from proteins to phosphoinositides to mRNAs. They play a very important role in cellular signalling within and between cells. They are key regulatory components in signal transduction pathways and their importance in the control of cellular signalling is well established. Furthermore, there are compelling reasons to believe that PTP inhibitors may serve as novel medicinal agents for the treatment of various diseases [205] including cancer as the protein tyrosine phosphatase activity has been correlated with a number of carcinomas including lung cancer [213215], colon and colorectal cancer [215,216], breast cancer, [217,218] brain and prostate cancer [218] and pancreatic cancer [219]. Protein Structure and Binding Site Definition The structure for this target contains 298 amino acid residues and was solved by Groves et. al by X ray diffraction at 2.1Å. PTP1B’s secondary structure includes 9 alpha helices and one main beta sheet composed of 8 strands as shown in figure x below. A structural feature present here that is highly conserved among Phosphotyrosine phosphatases is the catalytic, or PTP loop which is also known as the signature motif. This signature motif is a trademark characteristic for this superfamily and is the active site sequence C5XR. This PTP loop comprises 11 residues (shown in grey in figure x) but Cys 215 and Arg 221 are those most vital for catalysis. Another conserved loop, the recognition loop, is vital with regards to substrate recognition. Val 49 and Tyr 46 assist the substrate's insertion into catalytic site. Ser 216 of the PTP loop forms a hydrogen bond with the recognition loop, stabilizing the active site cleft. A third conserved loop is the WPD loop. On and near the WPD loop are key residues that function in PTP1B catalysis. Asp 181 and Gln 262 become especially important in the second part of the reaction. These structural features of PTP1B provide for the chemistry of dephosphorylation, detailed below. [221-222] The reaction starts when a phosphorylated tyrosine residue enters the deep, active site cleft of PTP1B molecule which lies just above the PTP loop. As mentioned earlier Tyr 46 and Val49 of the recognition loop facilitate this entry. Phosphotyrosine is an amphipathic molecule. The phosphorylated end of the tyrosine is polar but the phenol ring is non-polar and would normally be repelled from a polar catalytic site. Tyr46 and Val49 provide a non-polar pocket for the phenol ring of the phosphotyrosine substrate while the phosphorylated end is securely placed in the catalytic cleft. As soon as the substrate enters the catalytic site, a key conformational change occurs in the WPD loop. The loop closes over the phenyl ring of the tyrosine residue, holding it in place and further positioning it so that a subsequent nucleophilic attack may occur. At this same time, Asp 181 is moved in close to the tyrosine phosphate so it can act as an acid since it adds a proton (hydrogen) to the oxygen of tyrosine during the reaction and is therefore a crucial catalytic residue. Binding also occurs within the PTP loop; Arg 221 shifts to optimize its connection with the phosphate attached to the tyrosine residue. The slight shift of Arg 221 increases binding with Pro 180, Trp 179, and Phe 182. All of these interactions lead to a stabile, closed conformation for the WPD loop. Groves et al managed to crystallize the protein with a cyclic hexapeptide containing the phosphotyrosine analogue fluoromalonyl tyrosine (FOMT). The 5 residues of this hexapeptide were removed and only the phosphotyrosine was re-docked for this experiment. The interactions between the protein and this pTyr include hydrogen bonding (Cys 215, Ser 216, Ala 217, Ile 219, Gly 220, Gln 262), van der Waals bonds (Tyr 46, Asp 48) as well as anion-aromatic two planes interaction between the phosphotyrosine and Arg 221 and Gln 266. All of these residues as well as the ones mentioned above have been included in the definition of this binding site. This has resulted in a fairly ‘‘loose’’ binding site but as there are many residues involved in the whole process of tyrosine phosphorylation, if a particular ligand is able to inhibit their activity then it could prove to be a useful antagonist for this target. This strategy appears to have been successful as discussed in the results section of this chapter; the enrichment for this target is substantially above average (approximately 250 fold). Figure 5.33. PTP1B’s secondary structure includes 9 alpha helices (seen mostly on the left side of this illustration) and one main beta sheet composed of 8 strands (in the top and right side of this illustration). The PTP loop in the center of the structure is where the ligands bind (a cyclic hexapeptide containing the pTyr analogue fluoromalonyl tyrosine FOMT a.k.a. FLT) is docked in this instance, but for reasons of clarity only the FOMT is displayed) and is a hallmark for the PTP family of proteins and is crucial for this target’s catalytic activity. Figure 5.34. The surface display of the PTP1B site shows that it is exposed to the surface of the protein and as expected this is not a tight site as it needs to accommodate a few peptides for the tyrosine to be phosphorylated. A large number of residues are involved in the interactions with the pTyr and are labelled on the right graph. Although in this project it was generally desirable for reasons of CPU economy to have relatively small binding site definitions, this site has been expanded to the left of the ligand in order to include Asp 181 (coloured red because of its electronegativity in the left of the electronegative Arg 221). Asp 181 was targeted because it moves near the tyrosine phosphate during activation so it can act as an acid by adding a proton to the oxygen of tyrosine 3.2.2.7 The Smallpox Project Apart from the 12 cancer related proteins, the Screensaver platform was used to identify suitable molecules that could inhibit the type I topoisomerase protein of the vaccinia virus. The aim of inhibiting the function of this protein is to find a drug for smallpox. Smallpox, described as “the most deadly of diseases” by Jenner [225] was eliminated in 1980 as a result of a World Health Organisation (WHO) eradication program based primarily on vaccination [226]. Recent events, however, raise the possibility that the virus could be deliberately released and that the disease could make a comeback [227]. The level of immunity to smallpox in the general population is low partly because vaccination ceased in the UK in 1972 and so a large proportion of the population are unvaccinated and partly because re-vaccination is required to maintain immunity. 3.2.2.7.1 Vaccinia Topoisomerase Smallpox (variola) is a DNA virus which is related to the cowpox virus vaccinia used in vaccinations. Within the virus, the DNA is packaged as tightly wound supercoils, which are normally unwound by a type IB topoisomerase prior to viral replication. The variola topoisomerase I is very similar to that from vaccinia. Their amino acid sequences differ by only 2 amino acids (two glutamic acids are substituted by a glycine and a lysine at positions 47 and 159 respectively, and so the 2 proteins have 99.4% sequence identity). Moreover, an X-ray structure is available for the vaccinia topoisomerase [228], which raises the possibility of specifically designing compounds to inhibit the topoisomerase that could therefore be potential drugs against smallpox. Examination of the crystal structure has suggested two possible binding sites. The DNA binding site consists of the main catalytic residues, including Arg130, Lys167, Arg223, and His265, which are apparent in the Xray structure by Cheng et al.[228] The second site was identified by Charlton and Mortimer.[228] To fully describe this site, the loop (residues 129–137), which are missing in the X-ray structure were added and of the 12 possible conformations generated for this loop, 7 remained stable during molecular dynamics simulations and passed a number of tests. Figure 5.35. The DNA binding site for the vaccinia topoisomerase I is shown on the top two pictures. The protein is shown without the binding site on the left and with the binding site grid (in gray) on the right. The binding site energy grid method as implemented in Cerius2 is used for the evaluation of non bonded interactions between the rigid protein and the movable atoms from the flexible ligand. To visualise the second site identified by Charlton and Mortimer the protein needs to be rotated be 180o around its horizontal axis and this is shown in the bottom picture. 3.2.2.7.2 Human Topoisomerase In addition to searching for vaccinia topoisomerase inhibitors, a search for inhibitors of the human enzyme was initialised because such compounds may be potential anti-cancer drugs in their own right and because potential inhibitors of smallpox should ideally not interact with the human enzyme. Figure 5.36. A graphical display of the human topoisomerase complexed with a DNA double helix molecule. Here, topoisomerase in its closed form surround the DNA. In the left the snapshot was taken looking vertically on the axis of the double helix and on the right parallel to the DNA axis. The Smallpox Research Grip project was completed in 2004 after a total of 68,842 years of CPU time were utilised according to the Global MetaProcessor Statistics Service (www.grid.org/stats). In the first phase of the Screensaver Lifesaver project the set of 35 billion ligands was screened on the anthrax toxin using the THINK (http://www.treweren.com/) software to identify anthrax inhibitors. The results for both projects (anthrax, smallpox) have been delivered to the American Ministry of Defence. 3.2.2.8 Screensaver Lifesaver Phase II Results Once the docking and scoring of all ligands has completed, the most promising hits will be screened in the lab. The ligand scoring module runs ligscore, a fast, simple scoring function for predicting protein-ligand binding affinities in terms of pKi. So far we can report on the results from one of these targets, protein phosphatase 1B. The top 400 hits that can be bought or easily made were tested and 5% of these were found to be active; this compares to the 0.02% of actives that would have been expected if the compounds had been chosen randomly. This gives rise to a very encouraging enrichment factor of 250. 3.3 Conclusions NEED MORE ON THE CURRENT STATUS OF TESTING-INHIBOX?! REFERENCES 1. Begemann, J.H., Distributed Virtual High-Throughput Screening: A boon for drug discovery, Scientific Computing and Instrumentation, Tripos online industry publications (2004) 2. Waszkowycz, B., Perkins, T.D.J., Sykes, R.A. & Li J., Large-scale virtual screening for discovering leads in the postgenomic era, IBM systems Journal, 40, 2 (2001) 3. Richards, W. Graham, Virtual screening using grid computing: the screensaver project, Nature Reviews Drug Discovery, 1, 551-555 (2002) 4. H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne: The Protein Data Bank. Nucleic Acids Research, 28 pp. 235-242 (2000) 5. Venkatachalam, C.M., Jiang, X., Oldfield, T. & Waldman, M., LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites, Journal of Molecular Graphics and Modeling, 21 (4), 289-307 (2003) 6. Rojnuckarin, A., Livesay. A., Subramaniam, S., Bimolecular Reaction Simulation Using Weighted Ensemble Brownian Dynamics and the University of Houston Brownian Dynamics Program Biophys J, August 2000, p. 686-693, Vol. 79, No. 2 7. Dolle, R.E. Comprehensive survey of combinatiorial library synthesis: 2000., Journal of Combinatorial Chemistry, 3, 477-517 (2001) 8. Dolle, R. E. Comprehensive survey of combinatorial library synthesis: 1999. J. Comb. Chem. 2, 383–433 (2000). 9. Dolle, R. E. & Nelson, K. H. Jr. Comprehensive survey of combinatorial library synthesis: 1998. J. Comb. Chem. 1, 235–282 (1999). 10. Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. 23, 3-25 (1997) 11. Clark, D.E. & Pickett, S.D., Computational methods for the prediction of ‘druglikeness’, Drug Discovery Today, 5, 58-59 (2000) 12. Lesney Mark S., Finding new targets , Modern Drug Discovery, 4, 34-36 (2001) 13. Lindley, C., McCune, J.S., Thomason, T.E., Lauder, D., Sauls, A., Adkins, S. & Sawyer, W.T., Perception of chemotherapy side effects - Cancer versus noncancer patients, Cancer Practice, 7, (2) 59-65 (1999) 14. Carmeliet P, Jain RK: Angiogenesis in cancer and other diseases. Nature 2000, 407:249-257 15. Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanan, S. (2002) The protein kinase complement of the human genome. Science 298, 1912– 1934. 16. Davies, T.G., Bentley, J., Arris, C.E., Boyle, F.T., Curtin, N.J., Endicott, J.A., Gibson, A.E., Golding, B.T., Griffin, R.J., Hardcastle, I.R., Jewsbury, P., Johnson, L.N., Mesguiche, V., Newell, D.R., Noble, M.E.M., Tucker, J.A., Wang, L. & Whitfield, H.J. (2002) Structure-based design of a potent purinebased cyclin dependent kinase inhibitor. Nat. Struct. Biology 9, 745–749. 17. Franken, S. M., Scheidig, A. J., Krengel, U., Rensland, H., Lautwein, A., Geyer, M., Scheffzek, K., Goody, R. S., Kalbitzer, H. R., Pai, E. F., et al.: Threedimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras. Biochemistry 32, 8411 (1993) 18. Kurumbail, R. G., Stevens, A. M., Gierse, J. K., McDonald, J. J., Stegeman, R. A., Pak, J. Y., Gildehaus, D., Miyashiro, J. M., Penning, T. D., Seibert, K., Isakson, P. C., Stallings, W. C.: Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature 384, 644 (1996) 19. Wiesmann, C., Fuh, G., Christinger, H. W., Eigenbrot, C., Wells, J. A., de Vos, A. M.: Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell 91, 695 (1997) 20. Lah, M. S., Dixon, M. M., Pattridge, K. A., Stallings, W. C., Fee, J. A., Ludwig, M. L.: Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus. Biochemistry 34, 1646 (1995) 21. Hubbard, S. R.: Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J ,16, 5572 (1997) 22. Nagar, B., Bornmann, W., Pellicena, P., Schindler, T., Veach, D. R., Miller, W. T., Clarkson, B., Kuriyan, J.: Crystal Structures of the Kinase Domain of C-Abl in Complex with the Small Molecule Inhibitors Pd173955 and Imatinib (Sti-571) Cancer Res. 62, 4236 (2002) 23. Mohammadi, M., McMahon, G., Sun, L., Tang, C., Hirth, P., Yeh, B. K., Hubbard, S. R., Schlessinger, J.: Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955 (1997) 24. Lawrie, A. M., Noble, M. E., Tunnah, P., Brown, N. R., Johnson, L. N., Endicott, J. A.: Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol 4, 796 (1997) 25. Nassar, N.: The 2.2 A Crystal Structure of the Ras-Binding Domain of the Serine/Threonine Kinase C-Raf1 in Complex with RAP1A and a GTP Analogue Nature 375, 554 (1995) 26. Long, S. B., Casey, P. J., Beese, L. S.: The Basis for K-Ras4B Binding Specificity to Protein Farnesyltransferase Revealed by 2A Resolution Ternary Complex Structures Structure (London) 8, 209 (2000) 27. Mohammadi,M., McMahon, G., Sun, L., Tang, C., Hirth, P., Brian, K., Hubbard, S., Schlessinger, J., Structures of the Tyrosine Kinase Domain of Fibroblast Growth Factor Receptor in Complex with Inhibitors, Science, 276, 1997, 955961 28. M. Mohammadi, J. Schlessinger, S. R. Hubbard, Structure of the FGF receptor domain reveals a novel autoinhibitory mechanismCell 86, 577 (1996). 29. Groves, M. R., Yao, Z. J., Roller, P. P., Burke Jr., T. R., Barford, D.: Structural Basis for Inhibition of the Protein Tyrosine Phosphatase 1B by Phosphotyrosine Peptide Mimetics Biochemistry 37, 17773 (1998) 30. Pincus, M.R., Brandt-Rauf, P.W., Carty, R.P., Lubowsdy, J., Avitable,M., Gibson,K.D. and Scheraga, H.A. Proc. Natl Acad. Sci. USA, 84, 8375–8379. (1987) 31. Levitzki A., Signal-transduction therapy - A novel approach to disease management Eur. J. Biochem,226,1. , 1994 32. Pai A. F., Kengel U., Goody R. S., Kabsch W., Wittinghofer A., Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis, .EMBO J., 9, 2351. 1990 33. Barbacid M., "Ras gene". Annu. Rev. Biochem., 56, 779-827. 1987 34. Mayr B.; Schaffner G.; Reifinger M. K-ras mutations in canine pancreatic cancers The Veterinary Record, 19, vol. 153, no. 3, pp. 87-89(3) July 2003 35. de Vos AM, Tong L, Milburn MV, Matias PM, Jancarik J, Noguchi S, Nishimura S, Miura K, Ohtsuka E, Kim SH Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888-93 36. Milburn,M.V.; Tong,L.; deVos,A.M.; Brunger,A.; Yamaizumi,Z.; Nishimura,S.; Kim,S.H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990, 247 (4945): 939-945 37. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmuller L., Lautwein A., Schmitz F., Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science (Wash. DC), 277: 333-338, 1997 38. Masferrer, J. L., A. Koki, and K. Seibert. "COX-2 inhibitors. A new class of antiangiogenic agents." Ann.N.Y.Acad.Sci. 889 (1999): 84-86 39. Reddy BS, Hirose Y, Lubet R, Steele V, Kelloff G, Paulson S, Seibert K, Rao CV, Chemoprevention of colon cancer by specific cyclooxygenase-2 inhibitor, celecoxib, administered during different stages of carcinogenesis., Cancer Res. 2000 Jan 15;60(2):293-7 40. Jemal, A., Tiwari C., Murray, T., Ghafoo A., Samuels, A., Ward, A.., Feuer, E.J., Thun, M.J., Cancer statistics, 2004. CA Cancer J. Clin., 55: 8-31, 2004. 41. Thun M. J., Namboodiri M. M., Heath C. W., Jr. Aspirin use and reduced risk of fatal colon cancer. N. Engl. J. Med., 325: 1593-1596, 1991. 42. Giovannucci E., Egan K. M., Hunter D. J., Stampfer M. J., Colditz G. A., Willett W. C., Speizer F. E. Aspirin and the risk of colorectal cancer in women. N. Engl. J. Med., 333: 609-614, 1995. 43. Williams C, et al. The role of COX-2 in intestinal cancer. [Review] Ann N Y Acad Sci 1999;889:72-83 44. Watson AJ Chemopreventive effects of NSAIDs against colorectal cancer: regulation of apoptosis and mitosis by COX-1 and COX-2. Histol Histopathol 1998;13(2):591-7 45. Chan G, et al. Cyclooxygenase-2 expression is up-regulated in squamous cell carcinoma of the head and neck. Cancer Res 1999 Mar 1;59(5):991-4 46. Zimmermann KC, et al Cyclooxygenase-2 expression in human esophageal carcinoma. Cancer Res 1999 Jan 1;59(1):198-204 47. Wilson KT, et al. Increased expression of inducible nitric oxide synthase and cyclooxygenase-2 in Barrett's esophagus and associated adenocarcinomas, Cancer Res 1998 Jul 15;58(14):2929-34 48. Buckman SY, et al. COX-2 expression is induced by UVB exposure in human skin: implications for the development of skin cancer. Carcinogenesis 1998;19(5):723-9 49. Uefuji K, et al. Expression of cyclooxygenase-2 adenocarcinoma. J Surg Oncol 1998;69(3):168-72 50. J.K. Gierse, A single amino acid difference between cyclooxygenase-1 (COX1) and -2 (COX-2) reverses the selectivity of COX-2 specific inhibitors Journal of Biological Chemistry, 271, 15810 (1996). 51. D. Picot et al, Nature, The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1, 367, 243 (1994). 52. J.C. Otto and W.L. Smith, Prostaglandin endoperoxide synthases-1 and 2Journal of Biological Chemistry, 271, 9906 (1996). 53. A.S. Kalgutkar, Science, Covalent modification of cyclooxygenase-2 (COX-2) by 2-acetoxyphenyl alkyl sulfides, a new class of selective COX-2 inactivators280, 1268 (1998). 54. Hawkey CJ. COX-2 inhibitors. Lancet 1999;353(9149):307 55. Walsh K, et al. Angiogenic peptides in prostatic disease. BJU Int 1999; 84(9):1081-1083 56. Duque JL, et al. Plasma levels of vascular endothelial growth factor are increased in patients with metastatic prostate cancer. Urology. 1999; 54(3):5237 57. Sato K, et al. Expression of vascular endothelial growth factor gene and its receptor (flt-1) gene in urinary bladder cancer. Tohoku J Exp Med 1998;185(3):173-84 58. Langer I, et al. Expression of vascular endothelial growth factor (VEGF) and VEGF receptors in human neuroblastomas. Med Pediatr Oncol. 2000;34(6):386-393 59. Eggert A, et al. High-level expression of angiogenic factors is associated with advanced tumor stage in human neuroblastomas. Clin Cancer Res. 2000;6(5):1900-8. 60. Meister B, et al. Expression of vascular endothelial growth factor (VEGF) and its receptors in human neuroblastoma. Eur J Cancer. 1999 ;35(3):445-9 61. Maeda K, et al. Expression of vascular endothelial growth factor and thrombospondin-1 in colorectal carcinoma. Int J Mol Med 2000 Apr;5(4):373-8 62. Linder C, Linder S, Munck-Wikland E, Strander H. Independent expression of serum vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF) in patients with carcinoma and sarcoma. Anticancer Res. 1998 May-Jun;18(3B):2063-8. 63. Bikfalvi, A. and Bicknell, R. (2002) Recent advances in angiogenesis, antiangiogenesis and vascular targeting. Trends Pharmacol. Sci. 23(12), 576–582 64. Ferrara, N. (1999) Molecular and biological properties of vascular endothelial growth factor. J. Mol. Med. 77(7), 527–543. 65. Ferrara, N., Gerber, H. P., and LeCouter, J. The biology of VEGF and its receptors. (2003) Nat. Med. 9, 669-676 protein in gastric 66. Robinson, C. J., and Stringer, S. E. The splice variants of vascular endothelial growth factor (VEGF) and their receptors (2001) J. Cell Sci. 114, 853-865 67. Shibuya, M. Vascular endothelial growth factor receptor-2: its unique signaling and specific ligand, VEGF-E. (2003) Cancer Sci. 94, 751-756 68. Soker, S., Takashima, S., Miao, H. Q., Neufeld, G., and Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. (1998) Cell 92, 735-745 69. Liu, W. et al. (2002) Antiangiogenic therapy targeting factors that enhance endothelial cell survival. Semin. Oncol. 29(3 Suppl 11), 96–103. 70. Kanno, S. et al. (2000) Roles of two VEGF receptors, Flt-1 and KDR, in the signal transduction of VEGF effects in human vascular endothelial cells. Oncogene 19(17), 2138–2146. 71. Stacker, S.A. et al. (1999) A mutant form of vascular endothelial growth factor (VEGF) that lacks VEGF receptor-2 activation retains the ability to induce vascular permeability. J. Biol. Chem. 274(49), 34884–34892. 72. Ferrara N: VEGF and the quest Nat Rev Cancer 2002, 2:795-803. 73. Carmeliet P, Jain RK: Angiogenesis in cancer and other diseases. Nature 2000, 407:249-257 74. Ferrara, NRole of vascular endothelial growth factor in regulation of physiological angiogenesis . (2001) Am. J. Physiol. Cell Physiol. 280, C1358C1366 75. Brower, V. Tumor angiogenesis--new drugs on the block. (1999) Nat. Biotechnol. 17, 963-968 76. Shibuya, M. Structure and function of VEGF/VEGF-receptor system involved in angiogenesis. (2001) Cell Struct. Funct. 26, 25-35 77. Gille, H., Kowalski, J., Li, B., LeCouter, J., Moffat, B., Zioncheck, T. F., Pelletier, N., and Ferrara, N. Analysis of Biological Effects and Signaling Properties of Flt-1 (VEGFR-1) and KDR (VEGFR-2) (2001) J. Biol. Chem. 276, 3222-3230 78. Rahimi, N., Dayanir, V., and Lashkari, K. Receptor Chimeras Indicate That the Vascular Endothelial Growth Factor Receptor-1 (VEGFR-1) Modulates Mitogenic Activity of VEGFR-2 in Endothelial Cells* (2000) J. Biol. Chem. 275, 16986-16992 79. Bernatchez, P. N., Soker, S., and Sirois, M. G. (1999) J. Biol. Chem. 274, 31047-31054 80. Hiratsuka, S., Minowa, O., Kuno, J., Noda, T., and Shibuya, M. 9354Vascular Endothelial Growth Factor Effect on Endothelial Cell Proliferation, Migration, and Platelet-activating Factor (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 9349Synthesis Is Flk-1-dependent 81. Fong, G. H., Zhang, L., Bryce, D. M., and Peng, J. Increased hemangioblast commitment, not vascular disorganization, is the primary defect in flt-1 knockout mice (1999) Development 126, 3015-3025 82. Dunk, C., and Ahmed, A. Vascular Endothelial Growth Factor Receptor-2Mediated Mitogenesis Is Negatively Regulated by Vascular Endothelial Growth Factor Receptor-1 in Tumor Epithelial Cells (2001) Am. J. Pathol. 158, 265-273 for tumour angiogenesis factors. 83. Bader GD, Betel D, Hogue CW. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31(1):248-50 84. Lavelle, F., Michelson, A., and Dimitrijevic, L.: Biological Protection by Superoxide Dismutase, Biochem Biophys Res Commun 55, 350, 1973 85. Petkau, A., Chelack, W., Pleskach, S., Meeker, B., and Brady, C.: Radioprotection of Mice by Superoxide Dismutase, Biochem Biophys Res Commun 65, 886, 1975 Li, B., Fuh, G., Meng, G., Xin, X., 86. Gerritsen, M. E., Cunningham, B., and de Vos, A. M Receptor-selective Variants of Human Vascular Endothelial Growth Factor (2000) J. Biol. Chem. 275, 29823-29828 87. Takahashi H, Hattori S, Iwamatsu A, Takizawa H, Shibuya M, A Novel Snake Venom Vascular Endothelial Growth Factor (VEGF) Predominantly Induces Vascular Permeability through Preferential Signaling via VEGF Receptor-1 J. Biol. Chem., Vol. 279, Issue 44, 46304-46314, October 29, 2004 88. Fridovich, I.: Superoxide Radical and Superoxide Dismutase, Biochem Soc Trans 1, 48, 1973 89. Fridovich, I.: Superoxide Radical and Superoxide Dismutase, Acc Chem Res 5, 321, 1972 90. Paschen, W., and Weser, U.: Singlet Oxygen Decontaminating Activity of Erythrocuprein (Superoxide Dismutase), Biochim Biophys Acta 327, 217, 1973 91. Salin, M., and McCord, J.: Free Radicals and Inflammation. Protection of Phagocytosing Leukocytes by Superoxide Dismutase, J Clin Invest 56, 1319, 1975 92. Fee, J., and Teitelbaum, H.: Evidence that Superoxide Dismutase Plays a Role in Protecting Red Blood Cells Against Peroxidative Hemolysis, Biochem Biophys Res Commun 49, 150, 1972 93. Fee, J., Bergamini, R., and Briggs, R.: Observations on the Mechanism of the Oxygen Dialuric Acid Induced Hemolysis of Vitamin E-Deficient Rat Blood Cells and the Protective Roles of Catalase and Superoxide Dismutase, Arch Biochem Biophys 169, 160, 1975 94. Gregory, E., Goscin, S., and Fridovich, I.: Superoxide Dismutase and Oxygen Toxicity in a Eukaryote, J Appl Bacteriol 117, 456, 1974 95. Peeters-Joris, C., Vandevoorde, A., and Baudhuin, P.: Subcellular Localization of Superoxide Dismutase in Rat Liver, Biochem J 150, 31, 1975 96. Tyler, D.: Polarographic Assay and Intracellular Distribution of Superoxide Dismutase in Rat Liver, Biochem J 147, 493, 1975 97. Villafranca, J., Yost, F., and Fridovich, I.: Magnetic Resonance Studies of Manganese (III) and Iron (III) Superoxide Dismutases, J Biol Chem 249, 3532, 1974 98. Bannister, J., Bannister, W., and Wood, E.: Bovine Erythrocyte Cupro-zinc Protein. I. Isolation and General Characterization, Eur J Biochem 18, 178, 1971 99. Keele, B., McCord, J., and Fridovich, I.: Further Characterization of Bovine Superoxide Dismutase and the Isolation from Bovine Heart, J Biol Chem 246, 2875, 1971 100. Nyman, P.: A Modified Method for the Purification of Erythrocuprein, Biochim Biophys Acta 45, 387, 1960 101. Salvemini D, Riley DP, Nonpeptidyl mimetics of superoxide dismutase in clinical therapies for diseases. Cell Mol Life Sci. 2000 Oct;57(11):1489-92 102. Hileman E.A.; Achanta G.; Huang P. Superoxide dismutase: an emerging target for cancer therapeutics Expert Opinion on Therapeutic Targets, 1 December 2001, vol. 5, no. 6, pp. 697-710(14) 103. Rotilio, G., Calabrese, L., Mondovi, B., and Blumberg, W.: Electron Paramagnetic Resonance Studies of Cobalt-Copper Bovine Superoxide Dismutase, J Biol Chem 249, 3157, 1974 104. Rotilio, G., Morpurgo, L., Calabrese, L., and Mondovi, B.: On the Mechanism of Superoxide Dismutase. Reaction of the Bovine Enzyme with Hydrogen Peroxide and Ferrocyanide, Biochim Biophys Acta 302, 229, 1973 105. Rotilio, G., Morpurgo, L., Giovagnoli, C., and Calabrese, L.: Studies of the Metal Sites of Copper Proteins. Symmetry of Copper in Bovine Superoxide Dismutase and Its Functional Significance, Biochem 11, 2187, 1972 106. Forman, H., and Fridovich, I.: On the Stability of Bovine Superoxide Dismutase. The Effects of Metals, J Biol Chem 248, 2645, 1973 107. Calabrese, L., Polticelli, F., Capo, C., and Musci, G.: Involvement of the Copper in the Inhibition of Cu,Zn Superoxide Dismutase Activity at High pH, Free Rad., Free Radic Res Commun 12, 305, 1991 108. Forman, H., Evans, H., Hill, R., and Fridovich, I.: Histidine at the Active Site of Superoxide Dismutase, Biochem 12, 823, 1973 109. Rigo, A., Viglino, P., and Rotilio, G.: Kinetic Study of O2-Dismutation of Bovine Superoxide Dismutase. Evidence for Saturation of the Catalytic Sites by O2, Biochem Biophys Res Commun 63, 1013, 1975 110. Pisick, E., Jagadeesh, S., and Salgia R., Receptor Tyrosine Kinases and Inhibitors in Lung Cancer, The Scientific World JOURNAL (2004) 4, 589-604 111. Yu, H. and Rohan, T. (2000) Role of the insulin-like growth factor family in cancer development and progression. J. Natl. Cancer Inst. 92(18), 1472–1489. 112. Frittitta L, Vigneri R, Papa V, Goldfine ID, Grasso G, Trischitta V Breast Structural and functional studies of insulin receptors in human breast cancer. Cancer Res Treat. 1993;25(1):73-82. 113. Umayahara, Y. et al. (1994) Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J. Biol. Chem. 269(23), 16433– 16442. 114. Hebert, E., Herbelin, C., and Bougnoux, P. (1994) Analysis of the IGF-II receptor gene copy number in breast carcinoma. Br. J. Cancer 69(1), 120–124. 115. Kaiser, U. et al. (1993) Expression of insulin-like growth factor receptors I and II in normal human lung and in lung cancer. J. Cancer Res. Clin. Oncol. 119(11), 665–668. 116. Schardt, C. et al. (1993) Characterization of insulin-like growth factor II receptors in human small cell lung cancer cell lines. Exp. Cell Res. 204(1), 22– 29. 117. Rotsch, M. et al. (1992) Characterization of insulin-like growth factor I receptors and growth effects in human lung cancer cell lines. J. Cancer Res. Clin. Oncol. 118(7), 502–508. 118. Lukanova, A. et al. (2001) A prospective study of insulin-like growth factor-I, IGF-binding proteins-1, -2 and -3 and lung cancer risk in women. Int. J. Cancer 92(6), 888–892. 119. Wei L; Hubbard SR; Hendrickson WA; Ellis L (1995) Expression, characterization, and crystallization of the catalytic core of the human insulin receptor protein-tyrosine kinase domain. J Biol Chem 270: 8122-30. 120. Li, B. "c-Abl in Oxidative Stress, Aging and Cancer." Cell Cycle 4.2 (2005). 121. Kipreos,E.T. and Wang,J.Y.J. Cell cycle-regulated binding of c-Abl tyrosine kinase to DNA (1992) Science, 256, 382-385. 122. Dikstein,R., Heffetz,D., Ben-Neriah,Y. and Shaul,Y. c-abl has a sequencespecific enhancer binding activity (1992) Cell, 69, 751-757. 123. Jeffers M.; LaRochelle W.J.; Lichenstein H.S. Fibroblast growth factors in cancer: therapeutic possibilities Expert Opinion on Therapeutic Targets, 1 August 2002, vol. 6, no. 4, pp. 469-482(14) 124. 1)Faderl, S., Talpaz, M., Estrov, Z., and Kantarjian, H. M. Chronic Myelogenous Leukemia (1999) Ann. Intern. Med. 131, 207-219 125. 2)Sawyers, C. L. Chronic Myeloid Leukemia (1999) N. Engl. J. Med. 340, 13301340 126. Corbin, A. S., Buchdunger, E., Pascal, F., Druker B. J., of the Structural Basis of Specificity of Inhibition of the Abl Kinase by STI571 J. Biol. Chem., Vol. 277, Issue 35, 32214-32219, August 30, 2002 127. Wang J, Stockton DW, Ittmann M.The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate Clin Cancer Res. 2004 Sep 15;10(18 Pt 1):6169-78.’ 128. Kiyotsugu Yoshida,1 Kiyoshi Komatsu,2 Hong-Gang Wang,2 and Donald Kufe1 c-Abl Tyrosine Kinase Regulates the Human Rad9 Checkpoint Protein in Response to DNA Damage Molecular and Cellular Biology, May 2002, p. 32923300, Vol. 22, No. 10 129. David-Cordonnier Marie-Hélène +, Payet1 Dominique, D'Halluin Jean-Claude, Waring2, Michael J. Travers1 Andrew A. and Christian Bailly* The DNA-binding domain of human c-Abl tyrosine kinase promotes the interaction of a HMG chromosomal protein with DNA Nucleic Acids Research, Vol 27, Issue 11 22652270, 130. Wang,J.Y.J. Abl tyrosine kinase in signal transduction and cell-cycle regulation (1993) Curr. Opin. Genet. Dev., 3, 35-43. 131. Van Etten,R.A., Jackson,P. and Baltimore,D. The mouse type IV c-abl gene product is a nuclear protein, and activation of transforming ability is associated with cytoplasmic localization (1989) Cell, 58, 669-678. 132. Sawyers,C.L., McLaughlin,J., Goga,A., Havilik,M. and Witte,O. The nuclear tyrosine kinase c-Abl negatively regulates cell growth (1994) Cell, 77, 121-131. 133. Mattioni,T., Jackson,P.K., Bchini-Hooft van Huijsduijnen,O. and Picard,D. Cell cycle arrest by tyrosine kinase Abl involves altered early mitogenic response (1995) Oncogene, 10, 1325-1333. 134. Yuan,Z.-M., Huang,Y., Fan,M.-M., Sawyers,C., Kharbanda,S. and Kufe,D. Genotoxic drugs induce interaction of the c-Abl tyrosine kinase and the tumor suppressor protein p53(1996) J. Biol. Chem., 271, 26457-26460. 135. Schmandt, R. E., et al. "Expression of c-ABL, c-KIT, and platelet-derived growth factor receptor-beta in ovarian serous carcinoma and normal ovarian surface epithelium." Cancer 98.4 (2003): 758-64. 136. Jeffers,M.; Shimkets,R.; Prayaga,S.; Boldog,F.; Yang,M.; Burgess,C.; Fernandes,E.; Rittman,B.; Shimkets,J.; LaRochelle,W.J.; Lichenstein,H.S. Identification of a novel human fibroblast growth factor and characterization of its role in oncogenesis, Cancer Res., 61,7, 2001 137. Theillet, C., et al. "FGFRI and PLAT genes and DNA amplification at 8p12 in breast and ovarian cancers." Genes Chromosomes.Cancer 7.4 (1993): 219-26. 138. Adnane, J., et al. "BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers." Oncogene 6.4 (1991): 65963. 139. Wagner M, Lopez ME, Cahn M, Korc MSuppression of fibroblast growth factor receptor signaling inhibits pancreatic cancer growth in vitro and in vivo Gastroenterology. 1998 Apr;114(4):798-807 140. Kobrin MS, et al. Aberrant expression of type I fibroblast growth factor receptor in human pancreatic adenocarcinomas. Cancer Res 1993; 53(20):4741-4 141. Wang,F.; McKeehan,K.; Yu,C.; Ittmann,M.; McKeehan,W.L., Chronic activity of ectopic type 1 fibroblast growth factor receptor tyrosine kinase in prostate epithelium results in hyperplasia accompanied by intraepithelial neoplasia, Prostate, 58,1, 2004: 1-12 142. Giri D, Ropiquet F, Ittmann M. Alterations in expression of basic fibroblast growth factor (FGF) 2 and its receptor FGFR-1 in human prostate cancer. Clin Cancer Res. 1999 May;5(5):1063-71 143. Jin W, et al. Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res 2000 Mar 1;60(5):1221-4 144. Jin W, et al. Glioblastoma cell-specific expression of fibroblast growth factor receptor-1beta requires an intronic repressor of RNA splicing. Cancer Res 1999 Jan 15;59(2):316-9 145. Mohammadi,M., McMahon, G., Sun, L., Tang, C., Hirth, P., Brian, K., Hubbard, S. Schlessinger, J., Structures of the Tyrosine Kinase Domain of Fibroblast Growth Factor, Receptor in Complex with Inhibitors, Science, 276, 1997, 955961 146. K. A. Thomas, G. M. Smith, T. B. Thomas, R. J. Feldman, Proc. Natl. Acad. Sci. U.S.A. 79, 4843 (1982) 147. M. Mohammadi, J. Schlessinger, S. R. Hubbard, Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction Cell 86, 577 (1996). 148. Morgan, D.O. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu. Rev. Cell Dev. Biol. 13 (1997) 261-291. 149. Pavletich, N.P. (1999) Mechanisms of cyclin dependent regulations: structures of cdks, their cyclin activators, and cip and INK4 inhibitors. J. Mol. Biol. 287, 821–828. 150. Brown, N.R., Noble, M.E.M., Endicott, J.A. & Johnson, L.N. (1999) The structural basis for specificity of substrate and recruitment peptides for cyclindependent kinases. Nat. Cell Biol. 1, 438–443 151. Lowe, E.D., Tews, I., Cheng, K.-Y., Brown, N.R., Gul, S., Noble, M.E.M., Gamblin, S.J. & Johnson, L.N. (2002) Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry 41, 15625–15634. 152. Fischer, L., Endicott, J.A. & Meijer, L. (2002) Cyclin-dependent kinase inhibitors. Progress in Cell Cycle Research (Meijer, L., Jezequel, A. & Roberge, M., eds), pp. 235–248. Station Biologique de Roscoff. Life in Progress Editions, Roscoff. 153. Knockaert, M., Greengard, P. & Meijer, L. (2002) Pharmacological inhibitors of cyclin-dependent kinases, Trends Pharmacol. Sci. 23, 417–425 154. Mihara, M., et al. "Overexpression of CDK2 is a prognostic indicator of oral cancer progression." Jpn.J.Cancer Res. 92.3 (2001): 352-60. 155. De Moliner Erika, Brown Nick R. Johnson Louise N. Alternative binding modes of an inhibitor to two different kinases Eur. J. Biochem. 270, 3174-3181 (2003) 156. Lawrie, A.M., Noble, M.E.M., Tunnah, P.R., Brown, N.R., Johnson, L.N. & Endicott, J.A. (1997) Protein kinase inhibition by staurosporine: details of the molecular interaction determined by X-ray crystallographic analysis of a CDK2staurosporine complex. Nat. Struct. Biol. 4, 796–801. 157. Prade, L., Engh, R.A., Girod, A., Kinzel, V., Huber, R. & Bossmeyer, D. (1997) Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential. Structure 5, 1627–1637. 158. Lamers, M.B.A.C., Antson, A.A., Hiubbard, R.E., Scott, R.K. & Williams, D.H. (1999) Structure of the protein tyrosine kinase domain of C-terminal Src kinase (Csk) in complex with staurosporine. J. Mol. Biol. 285, 713–725. 159. Zhu, X., Kim, J.L., Newcomb, J.R., Rose, P.E., Sover, D.R., Toledo, L.M., Zhao, H. & Morgenstern, K.A. (1999) Structural analysis of the lymphocyte specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors. Structure 7, 651–661. 160. Jeffrey, P.D., Russo, A.A, Polyak, K., Gibbs, E., Hurwitz, J., Massague, J. and Pavletich, N.P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376 (1995) 313-320. 161. Otyepka, M., Køíž, Z. and Koèa, J. Dynamics and binding modes of free cdk2 and its two complexes with inhibitors studied by computer simulations. J. Biol. Struct. Dyn. 20 (2002) 141-154. 162. De Bondt, H.L., Rosenblatt, J., Jancarik, J., Jones, H.D., Morgan, D.O. and Kim, S.H. Crystal structure of cyclin-dependent kinase 2. Nature 363 (1993) 595-602. 163. Johnson, M. A. and Pinto, B. M. (2002) Molecular mimicry of carbohydrates by peptides. Aust. J. Chem. 55, 13–25 164. Avruch, J. et al. (2001) Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res 56, 127-155 165. Kolch, W. (2001) To be or not to be: a question of B-Raf? Trends Neurosci 24, 498-500 166. Kolch, W. (2000) Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J 351 Pt 2, 289305 167. Storm, S.M. et al. (1990) raf oncogenes in carcinogenesis. Crit Rev Oncog 2, 18 168. Alavi A, Hood JD, Frausto R, Stupack DG, Cheresh DA: Role of Raf in vascular protection from distinct apoptotic stimuli.Science 2003, 301:94-96 169. Hood JD, Bednarski M, Frausto R, Guccione S, Reisfeld RA, Xiang R, Cheresh A: Tumor regression by targeted gene delivery to the neovasculature. Science 2002, 296:2404-2407 170. Ruoslahti E: Antiangiogenics meet nanotechnology. Cancer Cell 2002, 2:97-98. 171. Pritchard, C. and McMahon, M. (1997) Raf revealed in life-or-death decisions. Nat Genet 16, 214-215 172. Kolch, W., Kotwaliwale, A., Vass, K., Janosch P., The role of Raf kinases in malignant transformation. Exp. Rev. Mol. Med. (2002)25 April, http://www.expertreviews.org/02004386h.htm 173. Sporn, M.B. (1996) The war on cancer. Lancet 347, 1377-1381 174. Morrison, D. K. "Mechanisms regulating Raf-1 activity in signal transduction pathways." Mol.Reprod.Dev. 42.4 (1995): 507-14 175. Cutler, R. E., Jr., et al. "Autoregulation of the Raf-1 serine/threonine kinase." Proc.Natl.Acad.Sci.U.S.A 95.16 (1998): 9214-19 176. Webb, C.P. et al. (1998) Signaling pathways in Ras-mediated tumorigenicity and metastasis. Proc Natl Acad Sci U S A 95, 8773-8778 177. Vojtek, A. B., Hollenberg, S. M., and Cooper, J. A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. (1993) Cell 74, 205-214 178. Chuang, E., Barnard, D., Hettich, L., Zhang, X.-f., Avruch, J., and Marshall, M. S. Critical binding and regulatory interactions between Ras and Raf occur through a small, stable N-terminal domain of Raf and specific Ras effector residues (1994) Mol. Cell. Biol. 14, 5318-5325 179. Ghosh, S., and Bell, R. M. Identification of discrete segments of human Raf-1 kinase critical for high affinity binding to Ha-Ras (1994) J. Biol. Chem. 269, 30785-30788 180. Emerson, S. D., Waugh, D. S., Scheffler, J. E., Tsao, K.-L., Prinzo, K. M., and Fry, D. C. Chemical shift assignments and folding topology of the Ras-binding domain of human Raf-1 as determined by heteronuclear three-dimensional NMR spectroscopy. (1994) Biochemistry 33, 7745-7752 181. Fabian, J. R., Vojtek, A. B., Cooper, J. A., and Morrison, D. K. A Single Amino Acid Change in Raf-1 Inhibits Ras Binding and Alters Raf-1 Function (1994) Proc. Natl. Acad. Sci. U. S. A. 91, 5982-5986 182. Sung, Y.-J., Carter, M., Zhong, J.-M., and Hwang, Y.-W. Mutagenesis of the Hras p21 at glycine-60 residue disrupts GTP-induced conformational change. (1995) Biochemistry 34, 3470-3477 183. Chow, Y.-H., Pulmiglia, K., Jun, T. H., Dent, P., Sturgill, T. W., and Jove, R. (1995) J. Functional Mapping of the N-terminal Regulatory Domain in the Human Raf-1 Protein Kinase Biol. Chem. 270, 14100-14106 184. Drugan JK., Khosravi-Far, R., White, MA., Der CJ., Sung YJ., Hwang, YW., Campbell SL., Ras Interaction with Two Distinct Binding Domains in Raf-1 May Be Required for Ras Transformation, Journal of Biological Chemistry,271, 1, 1996 pp. 233-237 185. Shirouzu, M., Koide, H., Fujita-Yoshigaki, J., Oshio, H., Toyama, Y., Yamsaki, K., Fuhrman, S. A., Villafranca, E., Kaziro, Y., and Yokoyama, S. Mutations that abolish the ability of Ha-Ras to associate with Raf-1. (1994) Oncogene 9, 21532157 186. Ahmed, S., Majeux, N. and Caflisch, A Hydrophobicity and functionality maps of farnesyltransferase. J. Mol. Graph. 19, 307-317, 380-307 (2001). 187. Zhang, H., Protein prenyltransferases, Handbook of Metalloproteins Vol. 3 John Wiley & Sons, Ltd, Chichester, 2004 pp. 37–48 188. Sebastian Maurer-Stroh, Stefan Washietl and Frank Eisenhaber, Protein prenyltransferases, Genome Biology 2003, 4:212 189. O'Regan RM, Khuri FR. Farnesyl transferase inhibitors: the next targeted therapies for breast cancer?Endocrine Related Cancer 2004; 11(2): 191-205 190. L. R. Kelland. "Farnesyl transferase inhibitors in the treatment of breast cancer". Expert Opin. Investig. Drugs 2003, 12, 413-21 191. W. C. M. Dempke. "Farnesyltransferase Inhibitors - a novel approach in the treatment of advanced pancreatic carcinomas". Anticancer Res. 2003, 23, 813818 192. J. E. Karp et al. "Current status of clinical trials of farnesyltransferase inhibitors" Curr. Opin. Oncol. 2001, 13, 470-476 193. M. Venet, D. End, P. Angibaud. "Farnesyl Protein Transferase Inhibitor ZARNESTRA® R115777 - History of a Discovery". Curr. Top. Med. Chem. 2003, 3, 1095-1102. 194. A. G. Taveras, P. Kirschmeier, C. M. Baum. "Sch-66336 (Sarasar®) and other Benzocycloheptapyridyl Farnesyl Protein Transferase Inhibitors: Discovery, Biology, and Clinical Observations". Curr. Top. Med. Chem. 2003, 3, 1103 1114 195. W. C. Rose et al. "Preclinical antitumor activity of BMS-214662, a highly apoptotic and novel farnesyltransferase inhibitor". Cancer Res. 2001, 61, 75077517 196. Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS: Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science 1997, 275:1800-1804 197. 21. Andres DA, Goldstein JL, Ho YK, Brown MS: Mutational analysis of alphasubunit of protein farnesyltransferase. Evidence for a catalytic role. J Biol Chem 1993, 268:1383-1390. 198. H. W. Park, S. R. Boduluri, J. F. Moomaw, P. J. Casey, L. S. Beese. "Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution". Science 1997, 275, 1800-1804. 199. S. B. Long, P. J. Casey, L. S. Beese. "The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures". Structure 2000, 8, 209-222. 200. Yokoyama K, McGeady P, Gelb MH: Mammalian protein geranylgeranyltransferase-I: substrate specificity, kinetic mechanism, metal requirements, and affinity labeling. Biochemistry 1995, 34:1344-1354. 201. Furfine ES, Leban JJ, Landavazo A, Moomaw JF, Casey PJ: Protein farnesyltransferase: kinetics of farnesyl pyrophosphate binding and product release. Biochemistry 1995, 34:6857-6862. 202. Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL:The Pfam protein families database. Nucleic Acids Res 2000, 28:263-266. 203. Wendt KU, Lenhart A, Schulz GE: The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J Mol Biol 1999, 286:175-187. 204. Long SB, Casey PJ, Beese LS: The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 Å resolution ternary complex structures. Structure Fold Des 2000, 8:209-222. 205. Stoker AW. Protein tyrosine phosphatases and signalling., J Endocrinol. 2005 Apr;185(1):19-33. 206. J. F. Hancock, K. Cadwallader, C. J. Marshall. "Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B)". EMBO J. 1991, 10, 641-646 207. Hunter, T. "Signaling--2000 and beyond." Cell 100.1 (2000): 113-27. 208. Neel, B. G. and N. K. Tonks. "Protein tyrosine phosphatases in signal transduction." Curr.Opin.Cell Biol. 9.2 (1997): 193-204. 209. Tonks, N. K. "PTP1B: from the sidelines to the front lines!" FEBS Lett. 546.1 (2003): 140-48. 210. Saltiel, A. R. and J. E. Pessin. "Insulin signaling pathways in time and space." Trends Cell Biol. 12.2 (2002): 65-71. 211. Andersen, J. N., Mortensen, O. H., Peters, G. H., Drake, P. G., Iversen, L. F., Olsen, O. H., Jansen, P. G., Andersen, H. S., Tonks, N. K., and Moller, N. P. (2001) Mol. Cell. Biol. 21, 7117-7136 212. Protein tyrosine phosphatase receptor-type O targeted by methylation in lung c.lancer. Cancer Biol.Ther. 3.12 (2004): 1194. 213. Motiwala, T., et al. "Protein tyrosine phosphatase receptor-type O (PTPRO) exhibits characteristics of a candidate tumor suppressor in human lung cancer." Proc.Natl.Acad.Sci.U.S.A 101.38 (2004): 13844-49. 214. Tsukamoto, T., et al. "Molecular analysis of the protein tyrosine phosphatase gamma gene in human lung cancer cell lines." Cancer Res. 52.12 (1992): 3506-09. 215. Kuo, M. L., T. S. Huang, and J. K. Lin. "Preferential requirement for protein tyrosine phosphatase activity in the 12-O-tetradecanoylphorbol-13-acetate- induced differentiation of human colon cancer cells." Biochem.Pharmacol. 50.8 (1995): 1217-22. 216. Villa, F., et al. "Crystal structure of the PTPL1/FAP-1 human tyrosine phosphatase mutated in colorectal cancer: evidence for a second phosphotyrosine substrate recognition pocket." J.Biol.Chem. 280.9 (2005): 8180-87 217. Li, J., et al. "PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer." Science 275.5308 (1997): 1943-47. 218. Mishra, S. and A. W. Hamburger. "Role of intracellular Ca2+ in the epidermal growth factor induced inhibition of protein tyrosine phosphatase activity in a breast cancer cell line." Biochem.Biophys.Res.Commun. 191.3 (1993): 106672. 219. Liebow, C., et al. "Somatostatin analogues inhibit growth of pancreatic cancer by stimulating tyrosine phosphatase." Proc.Natl.Acad.Sci.U.S.A 86.6 (1989): 2003-07 220. Andersen, J. N., Jansen, P. G., Echwald, S. M., Mortensen, O. H., Fukada, T., Del Vecchio, R., Tonks, N. K., and Moller, N. P. (2004) FASEB J. 18, 8-30 221. Jia, Z., Barford, d., Flint, A.J., and N.K.Tonks (1995). Structural Basis for Phosphotyrosine Peptide Recognition by PTP1B. Science 268: 1754-1758. 222. Pannifer A., Flint A., Tonks N., and Barford D.(1998). Visualization of the Cysteinyl-phosphate Intermediate of a Protein-tyrosine Phosphatase by X-ray Crystallography. The Journal of Biological Chemistry 273: 10454-10462 223. Silva, N. and Marcey, D., Protein Tyrosine http://www.clunet.edu/BioDev/omm/ptp1b/molmast.htm 14/1/2005) Phosphatase (Accessed 1B, on 224. Bartova, I., Otyepka, M., Koizi, Z., Koea, J., Cyclin-Dependent Protein Kinase-2 regulation by phosphorylation, a molecular dynamics study cell, Mol. Biol. Lett. 8. No. 2A. 2003, 574 225. Jenner, E., An Inquiry into the Causes and Effects of the Variolae Vaccinae; a Disease Discovered in some of the Western Counties of England, Particularly Gloucestershire, and Known by the Name of The Cow Pox'. (1798) 226. Barrett, S., Eradication versus control: the economics of global infectious disease policies, Bulletin of the World Health Organization, 82, 9, 683-688 (2004) 227. Stroh, M., Smallpox: could a deadly disease—declared eradicated on Earth— come back to haunt humans as a bioweapon? Life Science Infectious Viruses, Science World (2003) 228. Cheng, C., Kussi, P., Pavletich, N. & Shuman, S., Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases.’ Cell, 92, 841 (1998) 229. Mortimer T., Chalton M., Personnal communication Chapter 4 Biological background and importance of Gcoupled protein receptors and G-proteins 4.1 Introduction Of the several large families of receptors, by far the largest, most versatile and most ubiquitous is that of the seven-transmembrane (7TM) receptors also known as Guanine nucleotide binding protein coupled receptors (GPCRs). There are approximately 1000 genes identified to encode such receptors which represents more than 1% of the human genome [1-4], since the first receptors were cloned more than two decades ago [5]. These receptors regulate virtually all known physiological processes in mammals and eukaryotes in general including the sensation of exogenous stimuli, such as light, odors, and taste [1,6]. GPCRs are of pivotal importance in most cellular communication and signalling pathways and this is reflected in the plethora of drugs that target these receptors. The recent history has shown that Intervention at the level of GPCRs can produce excellent therapeutic targets. In excess of 50% of drugs on market target this superfamily of receptors with known native ligands [7]. The current total market for GPCR drugs such as antihistamines, AT1 antagonists, and beta-blockers is $60 billion. However still more than 600 of them remain orphan receptors for which the native ligands are unknown. The chemical diversity among the endogenous ligands is exceptional. They include biogenic amines, peptides, glycoproteins, lipids, nucleotides, ions, and proteases [5,7,8]. With current techniques available, identifying the native ligand for a GPCR in a process called deorphanization requires about 5 years and costs millions of dollars per receptor. This has become an apparent bottleneck for drug discovery that targets GPCRs. It also obstructs biomedical advance on many fronts. [8] Therefore it is not surprising that there has been a very significant shift in the research from “wet” to in silico chemistry and simulations for the study of the mechanistics of these vital proteins by means of homology modelling, virtual High Throughput screening and other computational techniques. 4.2 GPCR Structure and Function 4.2.1 General Structure and Receptor Classification Up to date the only GPCR structure that has been solved is that of the inactive (dark state) rhodopsin, first crystallized by Palchzewski et al in 2000 (PDB code 1F88) at 2.80 Å [9] and then in the following two years there were two more PDB entries for the same protein by Teller et al (2001, PDB code 1HZX) [10] and Okada et al (2002, PDB code 1L9H) [11] at resolutions of 2.80Å and 2.60Å respectively. GPCRs do not share any overall sequence homology (5,11). However the presence of seven transmembrane-spanning a-helical segments connected by alternating intracellular and extracellular loops is a structural feature common to all GPCRs. Another common feature is that the amino terminus (N- terminus) is located on the extracellular side and the carboxy terminus (C- terminus) on the intracellular side Fig. 1). However the terminal domains do show a significant amount of variation as can be seen in figure 1. Considerable sequence homology is nevertheless found within members of several subfamilies. There are three major subfamilies and these include the receptors related to the “light receptor” rhodopsin and the β 2-adrenergic receptor (family A, also referred to as class A), the receptors related to the glucagon receptor (family B), and the receptors related to the metabotropic neurotransmitter receptors (family C). Yeast pheromone receptors make up two minor unrelated subfamilies, family D (STE2 receptors) and family (STE3 receptors). In Dictyostelium Discoideum four different cAMP receptors constitute yet another minor, but unique, subfamily of GPCRs (family F) [5]. This chapter will focus mostly on family A receptors as members of this family only are discussed in the following chapters (rhodopsin, β2 adrenergic, bradykinin and chemokine receptors). The subfamily of rhodopsin/ β2 adrenergic receptor-like receptors (family A) is by far the largest and the most studied. Family A receptors can be subdivided further into six major subgroups as indicated in Figure 1. The overall homology among all type A receptors is relatively low however there is a significant number of highly conserved key residues which are indicated in figure 1 as white circles with black letters. The high degree of conservation among these key residues suggests that they have an essential role for either the structural or functional integrity of the receptors. The only residue that is conserved among all family A receptors is the arginine in the Asp-Arg-Tyr (DRY) motif at the cytoplasmic side of transmembrane segment TM3 [5, 11]. The other key motif commonly present in class A GPCRs is the Asn-Pro-any residue-any residue-Tyr (NPxxY) found in helix 7. In most family A receptors, a disulfide bridge (cysteines shown as black circles with white letters in figure 1) is connecting the second and third extracellular loop (EL-3) between the loops segment emerging of helix 3 and helix 5. In addition, a majority of the receptors have a palmitoylated cysteine in the carboxy-terminal tail causing formation of a putative intracellular loop. Family B receptors include approximately 20 different receptors for a variety of peptide hormones and neuropeptides, such as vasoactive intestinal peptide (VIP), calcitonin, PTH, and glucagon. Except for the disulfide bridge connecting the second (EL-2) and third extracellular loops (EL-3), family B receptors do not contain any of the structural features characterizing family A receptors (5). Notably, the important DRY motif is absent in family B receptors, and the prolines conserved among the family B receptors are distinct from the ones conserved among the family A receptors. The most prominent and distinctive characteristic of family B receptors is the large extracellular amino terminus (approximately 100 residues) containing several cysteines, which presumably form a network of disulfide bridges (16) as indicated in figure 1. Family C receptors are characterized by an exceptionally long amino terminus of 500–600 amino acid residues. Receptors from this family include the metabotropic glutamate and g-aminobutyricacid (GABA) receptors, the calcium receptors, the vomeronasal, mammalian pheromone receptors, and the recently identified putative taste receptors [5, 26]. Family C receptors have, similarly to family A and B receptors, two putative disulfide-forming cysteines in EL 2 and EL 3, respectively, but otherwise they do not share any conserved residues with family A and B receptors. The amino terminus of the metabotropic glutamate receptors displays remote sequence homology with bacterial periplasmic binding proteins (PBPs), especially with the leucine/isoleucine/valine binding protein [26]. The glutamate binding site has been proposed to be equivalent to the known amino acid binding site of PBPs; therefore, it is believed that the amino terminus of family C receptors contains the ligand-binding site [26, 27]. N terminus Family A. Rhodopsin/β2 adrenergic receptor-like Extracellular environment C C 2 H1 N H2 H5 H4 H3 P W Cell Membrane H7 H6 D P Family includes: 1 P N P Y C D H8 R Cytosol Y C terminus C Family B. Glucagon/VIP/ Calcitonin receptor-like N terminus C C C Extracellular environment C C C C C 2 1 1 H6 H7 P H1 H2 H3 Amine receptors (adrenergic, serotonin, muscarinic, histamine, dopamine) CCK, tachykinin, growth hormone secretagogues plus vertebrate opsins Invertebrate opsins and bradykinin Adenosine, cannobinoid, melanocortin and olfactory receptors Chemokine, eicosanoid, somatostatin, nucleotide,LH, opioid receptors + others Melatonin receptors + others P H4 H5 Cell Membrane W W Family includes: P Cytosol Calcitonin, CGRP and CRF receptors PTH and PTHrP receptors Glucagon, Glucagon-like peptide, GIP, GHRH, PACAP, VIP and secretin receptors Latrotoxin C terminus Family C. Metabotropic neurotransmitter/Calcium receptors N terminus Extracellular environment C C C P2 H1 H2 H3 H4 W H5 1 1 H6 H7 Cell Membrane Family includes: S T Metabotropic glutamate receptors Metabotropic GABA receptors Calcium receptors Vomeronasal pheromone receptors Taste receptors P P K P K N E Cytosol A C terminus Figure 4.1. The three main subfamilies of the GPCR proteins. The serpentine 7TM helix motif is found in all subfamilies. The most conserved residues are shown in black letters in white circles and the cysteine residues involved in disulphide bridges in the loops are shown in black circles with white letters. 4.2.2 Current GPCR crystal structures As mentioned earlier, the only available GPCR structures are that of the inactive state of bovine rhodopsin crystallized at 2.8 Å in 2000 [9,10] and 2.6Å [11] and these are commonly used as the template for some of the recent homology models published a selection of which is shown in references [28-35]. In the previous millennium, prior to the discovery of the structure of rhodopsin the homology models of GPCRs were based on the crystal structure of bacteriorhodopsin discovered by by electron diffraction in 1990 and by X-ray in 1995 both times at a resolution of 3.6Å. Some of these early homology models are discussed in references [36-40]. Bacteriorhodopsin, although a seven-spanning membrane protein found in a photosynthetic bacterium it does not activate any G protein and is therefore does not belong to the family of the GPCRs. The models which used bacteriorhodopsin as a template are now considered outdated as the salvation of rhodopsin showed that the relative positions and the angles of the helices in rhodopsin were not identical to those of bacteriorhodopsin. The crystal structures of rhodopsin and bacteriorhodopsin are shown in figure 4.2. Figure 4.2. The crystal structure of rhodopsin and bacteriorhodopsin. These two molecules have most commonly been used as templates for homology models of GPCRs. Rhodopsin on the left, has larger intracellular and extracellular loops as can be seen from the top panel (looking from the plane of the membrane). Bacteriorhodopsin lacks helix 8 which is responsible for the palmitoylation of the receptor. Bacteriorhodopsin has also got better defined α helical domains with fewer kinks and bends than bovine rhodopsin as can be seen from both the top and bottom panel (looking from outside of the cell, vertically towards the receptor) 4.2.2.1 Transmembrane domains The 7 α-helical domains that span the membrane are the most characteristic feature of GPCRs. All members of the rhodopsin-like (class A) GPCR family share common spatial structure of the transmembrane 7-α bundle which represents the most evolutionarily conserved part of these receptors. The 7 helices which are ordered in an anti-clockwise manner (figure 4.4) vary in size and range from 22 to 35 residues with H3 and H6 being the largest and H4 and H7 the smallest. As expected there is a high concentration of hydrophobic amino acids. Helices H1, H4, H6, and H7 are bent at proline residues which cause significant distortions especially in the extracellular end of H7. A proline residue is present in the middle of H5 (Pro215) however this helix is almost straight. There is a significant bend at Pro267 in H6. H7 exhibits irregular helicity, mainly around Lys 296 which is the residue where retinal is covalently bound. H2 also shows kinks because of the other ‘helix breaker’ residue, glycine (Gly 89 and Gly 90). In this region, because of this kink, H2 is closer to H3 than to H1, placing Gly90 to the key residue Glu113 [9] (homologous to Asp 113 in the β2 adrenergic receptor. This location of Gly90 is consistent with the previous studies showing that mutation of this residue by Asp causes night blindness (41), probably because of destabilization of the salt-bridge between Glu113 and the Schiff base [42-43]. The cytoplasmic terminal region is surrounded mostly by hydrophobic residues from H2 (Pro 71, Leu 72), IL-2 (Phe 148), H5 (Leu 226, Val 230), and H6 (Val 250, Met 253), forming the binding site for a G protein. H4 and H5 exhibit irregular helicity in the cytoplasmic region and at His 211, respectively. The phenolic ring of Tyr 223, which is also a highly conserved residue in GPCRs, partially covers the interhelical region between H5 and H6 near the lipid interface. Three basic residues, Lys 245, Lys 248, and Arg 252, located near the cytoplasmic end of H6, extend from the helical bundle, making this region of IL-3 highly basic. In H7, two phenyl rings of Phe 293 and Phe 294 interact with Leu 40 of H1 and Cys 264 of H6, respectively. This interaction with H6 is likely to be particularly important because it is facilitated by distortion of H6 in the region of Ile 263. H7 is considerably elongated in the region from Ala 295 to Tyr 301. This region includes Ala 299, whose peptide carbonyl can hydrogen bond with the side chains of Asn 55 in H1 and Asp 83 in H2. Details of this region are shown in Figure 4.3. A highly conserved NPXXY motif (in rhodopsin NPVIY) in GPCRs follows this region in a regular helical structure [9]. Figure 4.3. An accurate model of the secondary structure of rhodopsin before the crystal structure was published shows the distribution of the conserved residues and relative sizes of the different helices in rhodopsin. This figure is a modified version (for reasons of consistency) of the one published by Hardgrave and McDowell [44] and Palczewski [19]. The glycosylation and palmitoylation sites discussed later in this chapter have also been highlighted. Please note that this figure shows the intracellular domain on the top and the extracellular domain in the bottom opposite to the depictions in figures 4.1 and 4.2. 4.2.2.2 Intracellular and extracellular loops Although the role for the intracellular loops (ICLs) is fairly well defined as they are required for G-protein interaction and receptor regulation through kinases, arrestins, scaffolding proteins and other mediator proteins, a role for the 3 extracellular loops (ECLs) is not so apparent. An exception to this is ECL-2, which in the bacteriorhodopsin crystal structure forms contacts with 11-cis retinal by plunging deep into the TM bundle [9]. All of this may give the impression that ECL-1 and ECL-3 are peptide linkers incorporated into the GPCR structure merely to join functionally important TM helices together. However, there is now increasing evidence that this is not the case and that ECLs fulfil important functional roles within the GPCR architecture [45]. In the present study, we examine the functional significance of ECL-3, focusing primarily on Family A receptors. 4.2.3 Ligand binding Numerous studies have been carried out to identify domains involved in ligand binding to various subclasses of GPCRs, as this thesis involves small molecule binding family A receptors only their binding sites will be discussed here. The binding domains for endogenous ligands in this family of receptors, such as the site for the retinal photochromophore in rhodopsin and the binding site for catecholamines in the adrenergic receptors, are perhaps the best characterized. These are described in detail in references [46-49]. The photochromophore of rhodopsin and the opsins in general is 11cis-retinal which is unique among the endogenous ligands for GPCRs as it is the only ligand that is covalently bound to the receptor within a binding crevice formed by the transmembrane helices [50]. Through formation of a Schiff base, 11-cis-retinal is coupled to a lysine in TM 7 (Lys 296). The protonated Schiff base is paired with a glutamic acid (Glu 113) in the outer portion of TM 3 [51]. Additional interactions are found in TM 3 between the C9 group of retinal and Gly 121 [52], and between retinal and aromatic residues in the outer portion of TM 6 [53]. Upon exposure to light, 11-cis-retinal undergoes an isomerization to all-trans-retinal, which leads to formation of the metarhodopsin II state and thus receptor activation [50]. While all-trans-retinal behaves like the rhodopsin agonist, 11-cis-retinal behaves as an inverse agonist (i.e., an antagonist with negative intrinsic activity), keeping the receptor quiescent in the absence of light [12,54]. The binding sites for the classical “small-molecule” transmitters such as epinephrine, norepinephrine, acetylcholine are contained dopamine, in a serotonin, binding crevice histamine, and formed the by transmembrane helices. The residues involved in binding of agonists and antagonists to the β2- adrenergic receptor are found in TM 3, 5, 6, and 7 (see figure 4.4). The binding crevice is buried deep in the receptor molecule as demonstrated by spectroscopic analysis of the fluorescent antagonist carazolol bound to the β2-adrenergic receptor [55]. The energetically most important interaction is most likely a salt bridge between the charged amine of adrenergic ligands and the carboxylated side chain of Asp 113 in TM 3 [56] as frequently observed in the docking studies discussed in chapter 4. This aspartic acid is conserved among the biogenic amine receptors and is thought to interact also with the positively charged head group of dopamine [57], serotonin [58,59], histamine [60], and acetylcholine [61]. Additional key interactions of the agonists in the β2 -adrenergic receptor include hydrogen bonding between the hydroxyls of the catechol ring in epinephrine and two serines one a-helical turn apart in TM 5, Ser 204 and Ser 207 [60]. In TM 6, Phe 290 may stabilize the catechol ring [63] while recent evidence suggests that Asn 293 forms a hydrogen bind with the b-hydroxyl of epinephrine [64] In the case of the β2-adrenergic antagonists, which are structurally related to the endogenous agonists, evidence suggests that they share the Asp 113 ionic interaction with the agonists, but that other key interactions differ. For aryloxyalkylamine antagonists, such as alprenolol and propranolol, an asparagine in TM 7 (Asn 312) has been identified as a critical interaction point [65] (Fig. 3). Even though the majority of ligands for small-molecule transmitter receptors seem to bind deep within the binding crevice, there are indications that some antagonists, which show no structural relationship with their corresponding agonist, may partly interact with residues closer to the surface of the membrane. For example, α1-antagonists, such as phentolamine and WB4101, may interact with three residues in ECL 2 immediately adjacent to the top of TM 5 (66). H6 F S N H7 H5 S N D H1 H3 H2 H4 Figure 4.4. The complementarity of the binding cavities of receptors to their corresponding ligands is evident from the geometrical fit, the formation of intermolecular H-bonds, and from the clustering of receptor and ligand groups with similar polarity 4.2.4 Glycosylation It has been documented that GPCRs get glycosylated in one or more sites [18-21] in the extracellular domain, most commonly in arginine residues near the N-terminus (Figure 4.3). The purpose of this process is believed to be related with the correct distribution and trafficking of the receptors within the cell [18] although single point mutations at the glycosylation sites have been shown to produce functional receptors [22-23]. However, a single mutation in the glycosylation consensus sequence in human rhodopsin can result in retinitis pigmentosa, a blinding disease. Unlike palmitoylation, glycosylation is not thought to have an effect or be regulated by ligand binding of the receptor. 4.2.5 Palmitoylation As mentioned above, most of the class A GPCRs contain conserved cysteine residues at the cytoplasmic C-terminus, which represent putative palmitoylation sites. Palmitoylation of the GPCRs was confirmed experimentally by point mutagenesis and truncation analysis by Qanbar and Bouvier [70]. Palmitoylation has been shown to be a dynamic and agonist dependent modification as agonists have both shown to increase the incorporation of palpitate in the α2 and β2 adrenenoreceptors (71,79) and other receptors [80,81]. On the contrary, incorporation of the palmitate in the vasopressin V2 receptor was decreased upon the agonist stimulation (82). Some studies revealed that the GPCR palmitoylation may be involved in processing and membrane targeting of the receptors. Initial palmitoylation of the GPCRs occurs either in the endoplasmic reticulum-Golgi intermediate or in the early Golgi apparatus compartments [83] and appears to be important for the expression of the functional receptors on the cell surface. Studies have also correlated the replacement of the palmitoylation sites in several GPCRs with the receptor’s downstream signalling [81]. The nonpalmitoylated human somatostatin receptor type 5 displayed reduced coupling to adenylate cyclase [84], whereas mutation of a palmitoylation site in the β2adrenergic receptor impaired its interaction with the α subunit of the G protein [85, 86]. The cascade of events that leads to the desensitization of GPCRs is initiated by phosphorylation of the receptor by kinases described below, followed by interaction of the receptor with the β-arrestin and internalization. Palmitoylation of the GPCRs was proposed to play an important role in the regulation of receptor desensitization. Therefore, palmitoylation-deficient mutant of the β2 - adrenergic receptor was shown to be hyperphosphorylated at the basal level, and its phosphorylation did not increase upon the agonist stimulation [85]. 4.2.6 Phosphorylation GPCR activation by ligand binding usually leads to both to the initiation of signal transduction as explained in 4.3.1 and then to receptor internalization and desensitization (72-74). These two processes are adaptive mechanisms that prevent persistent receptor stimulation which could potentially lead to detrimental cellular effects. Internalization may also play a role in receptor resensitization. A critical first step in both GPCR internalization and desensitization is believed to be receptor phosphorylation by G proteinreceptor kinases and second-messenger-activated kinases. Phosphorylated receptors then bind regulatory proteins called arrestins, which inhibit further signaling by blocking receptor-G protein interaction (72-75). Arrestins also play a role in receptor internalization and some pathways of signal transduction by serving as adapters to localize essential proteins to the ligandactivated GPCR (76). These molecular events have been thoroughly characterized in a plethora of cell culture model systems and are assumed to occur similarly in vivo. However, the only direct evidence supporting the conclusion that activation leads to GPCR phosphorylation in intact animals comes from studies of rhodopsin. More specifically, the exposure of mice to light has been shown to stimulate the phosphorylation of retinal rhodopsin (77-78). To date, no such evidence has been provided with human material or with a ligandstimulated GPCR. Factors that have made the detection and study of agonist stimulated receptor phosphorylation notoriously difficult in vivo include the extremely low abundance of individual GPCR subtypes in tissues, the difficulty of GPCR purification, the absence of sufficiently sensitive methods for the detection of receptor phosphorylation, and, in the case of human tissues, the difficulty in getting tissue samples with and without exposure to an appropriate agonist [75] all of which are factors that have increased the demand for accurate computational modelling systems. 4.3 Receptor function and biological Significance Availability of reliable three-dimensional structures for GPCRs will greatly aid subtype specific structure-based drug design that would minimize the crossreactivity of drugs among other GPCRs. To partially solve the problem and to stimulate experiments to gain greater insight into GPCR structure and function, many research groups and pharma industry have utilized homologymodeling techniques based on the crystal structure of bovine rhodopsin (4). However a recent review by Baker, Sali and others (5) has shown that a homology model for a protein with 80 amino acids and with a sequence identity of less than 30% to the template crystal structure is unreliable. The sequence identity of bovine rhodopsin to many GPCRs is less than 20%, meaning a homology-based approach is unlikely to provide a reliable structure to be used for making predictions. However recent publications (iadanza) Despite these shortcomings, several groups have utilized a rhodopsin-based homology model with loose distance restraints obtained from mutation experiments to model the active site of many GPCR systems. Several other groups (6) have raised concern about the use of the bovine rhodopsin structure as a template for homology modeling since the bovine rhodopsin crystal structure with 11-cis retinal, an inverse agonist, could be in an inactive conformation. Hence the homology models based on the bovine rhodopsin crystal structure, with optimization using experimental information, are likely to provide good correlation for antagonists (since the rhodopsin structure is co- crystallized with an inverse agonist), but unlikely to capture the correct nature of the agonist binding site (6). Moreover it is not clear if antagonists preferentially bind only to active or inactive conformation of the receptor. It must be noted that for many GPCRs, such as the dopamine receptor, subtype specific agonist therapies are high desirable and the current level of structure prediction technology does not allow for the rational design of such ligands. 4. (a) Teeter M.M., Froimowitz M., Stec B., DuRand C.J. J Med Chem. 37, 2874-2888 (1994). (b) Trumpp-Kallmeyer S., Hoflack J., Bruinvels A., Hibert M. J Med Chem. 35, 3448-3462 (1992). (c) Neve K.A., Cumbay M.G., Thompson K.R., Yang R., Buck D.C., Watts V.J., DuRand C.J., Teeter M.M. Mol Pharmacol. 60, 373-381 (2001). (d) Varady J., Wu X., Fang X., Min J., Hu Z., Levant B., Wang S. J. Med. Chem. 46, 43774392 (2003). 5. Baker D., Sali A. Science 294, 93-96 (2001). 6. For a review, please see: Furse K.E., Lybrand T.P. J. Med. Chem. 46, 44504462 (2003). Numbering Schemes To facilitate comparison of residues between the large number of different receptors belonging to family A there is an obvious need to formulate and use a common numbering scheme. Currently, three different numbering schemes have been suggested but none of them have gained any wide acceptance. The Schwartz and Baldwin numbering schemes are, in principle, identical (13, 14). According to both schemes, the most conserved residue in each helix has been given a generic number describing their predicted relative position in a standard helix of 26 residues (13, 14).A given residue is then described by the helix in which it is located (I–VII) followed by a number indicating its position in the helix. For example, V.16 indicates residue number 16 in TM 5. However, the two numbering schemes are unfortunately incompatible with one another since they do not, except in helix 1, agree on the relative positioning of the conserved residues in the helices (13, 14). This problem is not apparent in the Ballesteros-Weinstein numbering scheme (15). In this scheme, the most conserved residue in each helix has been given the number 50, and each residue is numbered according to its position relative to this conserved residue. For example, 6.55 indicates a residue located in TM 6, five residues carboxy terminal to Pro6.50, the most conserved residue in helix 6 [17]. References: 1. Pierce, K.L. et al. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 2. Schoneberg T., Schulz A., Gudermann T. Rev. Physiol. Biochem. and Pharm., 144, 143-227 (2002). 3. Fredriksson, R. et al. (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 4. Venter, J.C et. al, The sequence of the human genome, Science 2001, 291, 1304-1351 5. Kolakowski Jr LF 1994 GCRDb: a G-protein-coupled receptor database. Receptors Channels 2:1–7 6. Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS 1999 Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96:541–551 7. Flower, D.R. Modelling G-protein-coupled receptors for drug desing. Biochem, Biophys Acta, 1999, 1422, 207-234 8. Feng YH, Sun Y, and Douglas JG. Gbg-Independent Constitutive Association of Gsa with SHP-1 and Angiotensin II Receptor AT2 Is Essential in AT2mediated Activation of SHP-1. Proc. Natl. Acad. Sci. USA 99:12049-12054 (2002) 9. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 (2000). 10. Teller, D.C., Okada, T., Behnke, C. A., Palczewski, K., Stenkamp, R. W., Advances in determination of a high resolution three-dimensional structure of rhodopsin, a model of g-protein-coupled receptors (gpcrs) Biochemistry 2001, 40, 7761-7772 11. Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E.M., Shichida, Y., Functional role of internal water molecules in rhodopsin revealed by X-ray Crystallography Proc Natl Acad Sci USA 2002, 99, 5982-5987 12. Gether U Uncovering Molecular Mechanisms Involved in Activation of G Protein-Coupled Receptors Endocrine Reviews 21, 2000 (1): 90-113 13. Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC 1992 Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol 11:1– 20 14. Schwartz TW, Gether U, Schambye HT, Hjorth SA 1995 Molecular mechanism of action of non-peptide ligands for peptide receptors. Curr Pharm Design 1:325–342 15. Baldwin JM, Schertler GF, Unger VM 1997 An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol 272:144–164 16. Ballesteros JA, Weinstein H 1995 Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein coupled receptors. Methods Neurosci 25:366–428 17. Ulrich CD, Holtmann M, Miller LJ 1998 Secretin and vasoactive intestinal peptide receptors: members of a unique family of G protein coupled receptors. Gastroentoerology 114:382–397 18. Watson. S., Artkinstall, S., The G-protein linked receptor facts book, Academic Press, London, 1994 19. Raymond J.R., Hinatowich M., Lefkowitz R.J., Caron, M.G., Adrenergic receptors. Models for regulation of signal transduction processes, Hypertension, 1990, 15, 119-131 20. Zhang Z., Austin S.C., Smyth, E.M., Glycosylation of the Human Prostacyclin Receptor: Role in Ligand Binding and Signal Transduction Molecular Pharmacology, Vol. 60, Issue 3, 480-487, September 2001 21. Michineau, S., Muller, L., Pizard, A., Alhenc-Gelas, F., Rajerison, R.M., Nlinked glycosylation of the human bradykinin B2 receptor is required for optimal cell-surface expression and coupling Biol. Chem., Vol. 385, pp. 49-57, January 2004 22. Bjoerkloef, K., Lundstroem. K., Abuin, L., Greasley P.J., Cotecchia, S. Coand post-translational Modification of the a1B-Adrenergic receptor: Effects on Receptor Expression and Function, Biochemistry, 2002, 41, 4281-4291 23. Hawtin S.R., Davies. A.L.R., Matthews G., Wheatley, M. Identification of the glycosylation sites usilised on the V1a vasopressin receptor and assessment of their role in receptor signalling and expression, Biochemical Journal 2001, 357, 73-81 24. Sung, C.H., Schneider, B.G., Agarwal, N., Papermaster, D.S., Nathans, J., Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa, Proceedings of the National Academy of Sciences of the USA, 1991, 88, 8840-8844 25. Sung, C.H., Davenport, C.M. Hennessey, J.C., Maumeneem I.H., Jacobson S.G., Heckenlively, J.R., Nowakowski, R., Fishman, G., Gouras P., Nathans J., Rhodopsin mutations in autosomal dominant retinitis pigmentosa, Proceedings of the National academy of Sciences of the USA, 1991, 88, 6481-6485 26. Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS 1999 Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96:541–551 27. O’Hara PJ, Sheppard PO, Thogersen H, Venezia D, Haldeman BA, McGrane V, Houamed KM, Thomsen C, Gilbert TL, Mulvihill ER 1993 The ligandbinding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron 11:41–52 28. Aburi, M. and P. E. Smith. "Modeling and simulation of the human delta opioid receptor." Protein Sci. 13.8 (2004): 1997-2008. 29. Bissantz, C., et al. "Protein-based virtual screening of chemical databases. II. Are homology models of G-Protein Coupled Receptors suitable targets?" Proteins 50.1 (2003): 5-25. 30. Broer, B. M., M. Gurrath, and H. D. Holtje. "Molecular modelling studies on the ORL1-receptor and ORL1-agonists." J.Comput.Aided Mol.Des 17.11 (2003): 739-54. 31. Gouldson, P. R., et al. "Toward the active conformations of rhodopsin and the beta2-adrenergic receptor." Proteins 56.1 (2004): 67-84. 32. Montero, C., et al. "Homology models of the cannabinoid CB1 and CB2 receptors. A docking analysis study." Eur.J.Med.Chem. 40.1 (2005): 75-83. 33. Moro, S., A. H. Li, and K. A. Jacobson. "Molecular modeling studies of human A3 adenosine antagonists: structural homology and receptor docking." J.Chem.Inf.Comput.Sci. 38.6 (1998): 1239-48. 34. Neve, K. A., et al. "Modeling and mutational analysis of a putative sodiumbinding pocket on the dopamine D2 receptor." Mol.Pharmacol. 60.2 (2001): 373-81. 35. Prusis, P., et al. "Modeling of the three-dimensional structure of the human melanocortin 1 receptor, using an automated method and docking of a rigid cyclic melanocyte-stimulating hormone core peptide." J.Mol.Graph.Model. 15.5 (1997): 307-17, 334. 36. Cronet, P., C. Sander, and G. Vriend. "Modeling of transmembrane seven helix bundles." Protein Eng 6.1 (1993): 59-64. 37. Grotzinger, J., et al. "A model for the C5a receptor and for its interaction with the ligand [corrected]." Protein Eng 4.7 (1991): 767-71. 38. Homan, E. J., H. V. Wikstrom, and C. J. Grol. "Molecular modeling of the dopamine D2 and serotonin 5-HT1A receptor binding modes of the enantiomers of 5-OMe-BPAT." Bioorg.Med.Chem. 7.9 (1999): 1805-20. 39. Kyle, D. J., et al. "A proposed model of bradykinin bound to the rat B2 receptor and its utility for drug design." J.Med.Chem. 37.9 (1994): 1347-54. 40. Trumpp-Kallmeyer, S., et al. "Modeling of G-protein-coupled receptors: application to dopamine, adrenaline, serotonin, acetylcholine, and mammalian opsin receptors." J.Med.Chem. 35.19 (1992): 3448-62. 41. V. R. Rao, G. B. Cohen, D. D. Oprian, Characterization of the mutant visual pigment responsible for congenital night blindness: a biochemical and Fourier-transform infrared spectroscopy study Nature 367, 639 (1994). 42. T. P. Sakmar, R. R. Franke, H. G. Khorana, Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsinProc. Natl.Acad. Sci. U.S.A. 86, 8309 (1989). 43. E. A. Zhukovsky and D. D. Oprian, Effect of carboxylic acid side chains on the absorption maximum of visual pigmentsScience 246, 928 (1989). 44. Hargrave, P. A. and J. H. McDowell. "Rhodopsin and phototransduction: a model system for G protein-linked receptors." FASEB J. 6.6 (1992): 2323-31. 45. Z. Lawson and M. Wheatley1 The third extracellular loop of G-protein-coupled receptors: more than just a linker between two important transmembrane helices Biochemical Society Transactions (2004) Volume 32, part 6,10481051 46. 16 Kobilka B 1992 Adrenergic receptors as models for G protein-coupled receptors. Annu Rev Neurosci 15:87–114 47. 17Savarese TM, Fraser CM 1992 In vitro mutagenesis and the search for structure-function relationships among G protein-coupled receptors. Biochem J 283:1–19 48. 19Strader CD, Fong TM, Graziano MP, Tota MR 1995 The family of Gprotein-coupled receptors. FASEB J 9:745–754 49. 21. Ji TH, Grossmann M, Ji I 1998 G protein-coupled receptors. I. Diversity of receptor-ligand interactions. J Biol Chem 273:17299–17302 50. 66 Sakmar TP 1998 Rhodopsin: a prototypical G protein-coupled receptor. Prog Nucleic Acid Res Mol Biol 59:1–34 51. 67 Zhukovsky EA, Robinson PR, Oprian DD 1992 Changing the location of the Schiff base counterion in rhodopsin. Biochemistry 31:10400–10405 52. 68 Han M, Groesbeek M, Sakmar TP, Smith SO 1997 The C9 methyl group of retinal interacts with glycine-121 in rhodopsin. Proc Natl Acad Sci USA 94:13442–13447 53. 69 Nakayama TA, Khorana HG 1991 Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem 266:4269–4275 54. 70 Han M, Lou J, Nakanishi K, Sakmar TP, Smith SO 1997 Partial agonist activity of 11-cis-retinal in rhodopsin mutants. J Biol Chem 272:23081–23085 55. 71Tota RT, Strader CD 1990 Characterization of the binding domain of the beta-adrenergic receptor with the fluorescent antagonist carazolol. J Biol Chem 265:16891–16897 56. 72Strader CD, Gaffney T, Sugg EE, Candelore MR, Keys R, Patchett AA, Dixon RAF 1991 Allele-specific activation of genetically engineered receptors. J Biol Chem 266:5–8 57. 73Mansour A, Meng F, Meador-Woodruff JH, Taylor LP, Civelli O, Akil H 1992 Site-directed mutagenesis of the human dopamine D2 receptor. Eur J Pharmacol 227:205–214 58. 74Wang CD, Gallaher TK, Shih JC 1993 Site-directed mutagenesis of the serotonin 5-hydroxytrypamine2 receptor: identification of amino acids necessary for ligand binding and receptor activation. Mol Pharmacol 43:931– 940 59. 75Ho BY, Karschin A, Branchek T, Davidson N, Lester HA 1992 The role of conserved aspartate and serine residues in ligand binding and in function of the 5-HT1A receptor: a site-directed mutation study. FEBS Lett 312:259–262 60. 76Gantz I, DelValle J, Wang LD, Tashiro T, Munzert G, Guo YJ, Konda Y, Yamada T 1992 Molecular basis for the interaction of histamine with the histamine H2 receptor. J Biol Chem 267:20840–20843 61. 77Spalding TA, Birdsall NJ, Curtis CA, Hulme EC 1994 Acetylcholine mustard labels the binding site aspartate in muscarinic acetylcholine receptors. J Biol Chem 269:4092–4097 62. Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RAF 1989 Identification of two serine residues involved in agonist activation of the beta adrenergic receptor. J Biol Chem 264:13572–13578 63. Tota RT, Candelore MR, Dixon RAF, Strader CD 1990 Biophysical and genetic analysis of the ligand-binding site of the beta-adrenoceptor. Trends Pharmacol Sci 12:4–6 64. Wieland K, Zuurmond HM, Krasel C, Ijzerman AP, Lohse MJ 1996 Involvement of Asn-293 in stereospecific agonist recognition and in activation of the ß2-adrenergic receptor. Proc Natl Acad Sci USA 93:9276–9281 65. Suryanarayana S, Daunt DA, Von Zastrow M, Kobilka BK 1991 A point mutation in the seventh hydrophobic domain of the alfa-2 adrenergic receptor increases its affinity for a family of β-receptor antagonists. J Biol Chem 266:15488–15492 66. Zhao MM, Hwa J, Perez DM 1996 Identification of critical extracellular loop residues involved in α1-adrenergic receptor subtype-selective antagonist binding. Mol Pharmacol 50:1118–1126 67. Henderson, R., Baldwin, J. M., Ceska, T. A., Zemlin, F., Beckmann, E., Downing, K. H.: Model for the structure of bacteriorhodopsin based on highresolution electron cryo-microscopy. J Mol Biol 213 pp. 899 (1990) 68. Grigorieff, N., Ceska, T. A., Downing, K. H., Baldwin, J. M., Henderson, R.: Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol 259 pp. 393 (1996) 69. Conn PJ, Pin JP 1997 Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol 37:205–23 70. Qanbar, R., and Bouvier, M. (2003). Role of palmitoylation/depalmitoylation reactions in G-protein-coupled receptor function. Pharmacol Ther 97, 1-33. 71. Mouillac, B., Caron, M., Bonin, H., Dennis, M., and Bouvier, M. (1992). Agonist modulated palmitoylation of beta 2-adrenergic receptor in Sf9 cells. J Biol Chem 267, 21733-21737. 72. Ferguson SS, Caron MG 1998 G protein-coupled receptor adaptation mechanisms. Semin Cell Dev Biol 9:119–127 73. Pitcher JA, Freedman NJ, Lefkowitz RJ 1998 G protein-coupled receptor kinases. Annu Rev Biochem 67:653–692 74. Krupnick JG, Benovic JL 1998 The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol 38:289– 319 75. Liu, Q., Reubi, J-C., Wang, Y., Knoll B.J., Schonbrunn A., In Vivo Phosphorylation of the Somatostatin 2A Receptor in Human Tumors, The Journal of Clinical Endocrinology & Metabolism Vol. 88, 2003, No. 12 60736079 76. Miller WE, Lefkowitz RJ 2001 Expanding roles for beta-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr Opin Cell Biol 13:139– 145 77. Ohguro H, Van Hooser JP, Milam AH, Palczewski K 1995 Rhodopsin phosphorylation and dephosphorylation in vivo. J Biol Chem 270:14259– 14262 78. Kennedy M. J, Lee KA, Niemi GA, Craven KB, Garwin GG, Saari JC, Hurley JB 2001 Multiple phosphorylation of rhodopsin and the in vivo chemistry underlying rod photoreceptor dark adaptation. Neuron 31:87–101 79. Kennedy, M. E., and Limbird, L. E. (1994). Palmitoylation of the alpha 2Aadrenergic receptor. Analysis of the sequence requirements for and the dynamic properties of alpha 2A-adrenergic receptor palmitoylation. J Biol Chem 269, 31915-31922. 80. Ponimaskin, E. G., Schmidt, M. F., Heine, M., Bickmeyer, U., and Richter, D. W. (2001).5-Hydroxytryptamine 4(a) receptor expressed in Sf9 cells is palmitoylated in an agonist-dependent manner. Biochem J 353, 627-634. 81. Hayashi, M. K., and Haga, T. (1997). Palmitoylation of muscarinic acetylcholine receptor m2 subtypes: reduction in their ability to activate G proteins by mutation of a putative palmitoylation site, cysteine 457, in the carboxyl-terminal tail. Arch Biochem Biophys40, 376-382. 82. Sadeghi, H. M., Innamorati, G., Dagarag, M., and Birnbaumer, M. (1997). Palmitoylation of the V2 vasopressin receptor. Mol Pharmacol 52, 21-29. 83. Bradbury, F. A., Kawate, N., Foster, C. M., and Menon, K. M. (1997). Posttranslational processing in the Golgi plays a critical role in the trafficking of the luteinizing hormone/human chorionic gonadotropin receptor to the cell surface. J Biol Chem 272, 5921-5926. 84. Hukovic, N., Panetta, R., Kumar, U., Rocheville, M., and Patel, Y. C. (1998). The cytoplasmic tail of the human somatostatin receptor type 5 is crucial for interaction with adenylyl cyclase and in mediating desensitization and internalization. J Biol Chem 273, 21416-21422. 85. Moffett, S., Mouillac, B., Bonin, H., and Bouvier, M. (1993). Altered phosphorylation and desensitization patterns of a human beta 2-adrenergic receptor lacking the palmitoylated Cys341. Embo J 12, 349-356. 86. O'Dowd, B. F., Hnatowich, M., Caron, M. G., Lefkowitz, R. J., and Bouvier, M. (1989). Palmitoylation of the human beta 2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J Biol Chem 264, 7564-7569. Chapter 5 Towards the active conformations of rhodopsin and the β2-adrenergic receptor 5.1 Introduction Members of the Integral membrane G-protein coupled receptor (GPCR) family mediate many vital cellular signalling processes in eucarya. The landmark determination of the X-ray structure of a GPCR, rhodopsin [1], has given rise to new opportunities, e.g. for constructing models of other GPCRs by homology modelling. However, the only crystal structure to date is that of an inactive (dark state) form of the receptor. Consequently, we have systematically separated a wealth of additional structural data into data relevant to the inactive form and that relevant to the active form, with a view to employing this data to study the active form of the receptor. The structural data includes site-directed spin labelling (SDSL) experiments which involved cross-linking of cysteine residues with nitroxides, followed by EPR spectroscopy to determine interatomic distances; this technique has given extensive data on the structural changes that occur during rhodopsin photo-activation, most notably the movement of transmembrane helices upon activation[2-11]. The introduction of zinc binding sites was used to demonstrate unequivocally that the transmembrane helices of the NK1 tachykinin receptor were orientated in an anticlockwise fashion[12]; engineered zinc binding sites have also been used to study both inactive and active receptor conformations [13-18]. The substituted cysteine accessibly method (SCAM) has proved a powerful technique for determining the transmembrane residues that form the interior polar cavity of the dopamine D2 receptor [19-22]; SCAM has also been used to monitor conformational changes that occur in transmembrane helix 6 (TM6) of a constitutively active β2 -adrenergic ( 2-AR) receptor [23]. Site-directed crosslinking (SDCL) studies have provided the most illuminating structural details for the active receptor conformation; analysis of disulfide bond formation for native and introduced cysteine residues has demonstrated that large structural changes may occur during receptor activation[24-27]. An alternative cross-linking method that has been used for rhodopsin involves incorporating photo-active groups into ligand structures to provide details on ligand receptor interactions [28-31]. However, care must be taken when using this additional structural data, particularly with regard to whether the data correspond to active or inactive conformations. Thus for site-directed mutagenesis data, the effects of mutations on inverse agonists/antagonists and agonists must be treated separately. To date, many models have incorporated site-directed mutagenesis data for all classes of ligands to generate one receptor model. This results in an averaging of the models to a state that represents neither an active nor an inactive receptor, even if simulations are then performed in the presence of agonists or antagonists. Relatively few models have been built with at least two receptor conformations in mind [32,33] and this is one plausible origin for the discrepancies that occur between models and between models and experiment [34-38]. In order to address this problem, restrained molecular dynamics (MD) has been used to generate active (and inactive) conformational models of the human β2-AR receptor and rhodopsin with a view to using these models in further biochemical experiments on the molecular mechanisms that govern ligand interactions and receptor activation. 5.2 Computational Methods 5.2.1 Construction of the initial receptor models The model of the β 2 -AR receptor was built from the 2.8 Å rhodopsin structure [39] (pdb code 1F88) using the correspondences defined in the profile-based multiple sequence alignment of the β 2-AR and rhodopsin receptor families. (The RMS difference between this structure and the more recent 1HLX and 1IL9 structures are small.) Here and elsewhere the residues are identified by the sequence number in bovine rhodopsin or the rat 2-AR receptor, and by their GPCRDB40 global number (gn) [41] (see Table I). The undefined region of IL3 (Gln236-Glu239) in rhodopsin was added by searching for the best insert available in a refined protein structural database based on criteria such as gap closure distance and backbone atom position; sequence conservation was taken into account through user inspection of potential loop conformations. The large IL3 (Lys232-Leu258) and C-terminus (Ser347-Leu383) of the 2-AR receptor were not included in the receptor model. PLIM [42] was used to add 580 explicit water molecules to solvate the extracellular loops, intracellular loops and the transmembrane helical bundle cavity through an iterative process whereby water molecules were added in batches of 10, followed by energy minimisation employing a 15 Å cut off and a 30 step angle for finding the best hydrogen bond geometries. TABLE 5.1. The X-ray, sequence and generic numbers (GPCRDB and Ballesteros) for the maximum extent of helix ends, as determined by DSSP. Where the helix ends differ between the 4 different models, the numbers giving the longest helix are recorded. Rhodopsin 2 -AR TM X-ray X-ray Generic GPCRDB Ballesteros 1 34-64 30-60 109-139 1.28-1.58 2 71-100 67-96 212-241 2.38-2.67 3 106-140 102-136 311-345 3.21-3.55 4 150-175 147-172 409-434 4.39-4.64 5 199-230 195-226 504-535 5.34-5.65 6 246-277 267-298 599-630 6.29-6.60 7 285-309 305-329 712-736 7.32-7.56 Several approaches for modelling the membrane environment are possible. One approach is to carry out full lipid simulations on fully hydrated bilayers (or bilayer mimics) as the hydrophobic lipid chains provide packing forces and the solvent region provides dielectric screening. However, these methods are generally used for structural studies on channels that undergo limited conformational changes, e.g. see references [43-47]. Few membrane simulation studies have seriously addressed the true composition of the lipid but rather deal with ideal systems. Moreover, for systems undergoing large conformational changes, it is more usual to omit the solvent and the lipid as the lipid can hinder motion, reduce conformational sampling (and therefore increase errors) and introduce dielectric screening via a distance dependent dielectric. Indeed, in a rare study of GPCR activation in a fully hydrated bilayer, Rohrig and Rothlisberger admit that the simulations do not sufficiently model the receptor movement[49]. In contrast, the stochastic boundary molecular dynamics study of aquaporin [49] included more discussion of protein structural changes than related hydrated bilayer studies [45]. Consequently, the use of simplified methods is the usual approach for implementing NMR restraints [50,51] and by analogy is a natural choice for the simulations described here. Studies on the effect of mutations on redox potentials have shown that a distance dependent dielectric constant can give reliable predictions [52], and while the reported optimum screening was provided by an exponential function, here we have opted for a 1/r function since some screening is implemented explicitly through the incorporation of the explicit water molecules. Moreover, McCammon has shown that a few layers of explicit water molecules are an important adduct to a continuum treatment of a membrane environment [53] and while McCammon’s Poisson Boltzmann continuum treatment is more sophisticated than that used here, the combination of continuum plus explicit water is nevertheless well founded. The 11-cis and all trans conformations of retinal were docked into the crystal structure using interactive molecular graphics, thus generating two initial rhodopsin models. The solvated receptor models were subjected to MD simulations involving a 100 ps heating stage from 0 K to 310 K, a further 200 ps at 310 K and then a conjugate gradient minimisation to yield the initial solvated receptor models. Restraints for inactive and active receptor conformations. Restraints for residues and atoms relevant to inactive and active receptor conformations were calculated, primarily from SDSL, SCAM and engineered zinc binding data and are given in Tables II and III respectively; the Tables also show the actual distances observed in the rhodopsin crystal structure. The restraints from the SDCL are particularly important since the structure of a disulfide bond is well defined and is chemically compatible with protein structures in general. Moreover, zinc is coordinated by His in a welldefined manner [54] but distance restraints for C carbon positions do not necessarily provide sufficient accuracy to form the binding site. Supplementary restraints were required for C and imidazole nitrogen distances to produce a valid histidine zinc binding site. The residues were therefore mutated to the most appropriate His rotamer in the receptor model to ensure that the relative orientations of the residues were correct for coordination with zinc. The restraints used [55] were that C carbons should be separated by no more than 11 Å, and the N-N distance should be approximately 6 Å. To allow proper inclusion of the SDSL data, a nitroxidesubstuted cysteine residue was constructed (Fig. 1.) so that the relevant r(OO) distance restraint from Tables II and III could be included explicitly in 100 ps simulations of receptors each containing a nitroxide pair. In one of the 2AR receptors containing a nitroxide at position gn601, the Arg131(gn330) Glu268(gn600) interaction was broken and the restraint was more readily satisfied than in the other simulations. Consequently, the 100 ps nitroxide simulations were repeated with the salt bridge broken by an E268A mutation. From these simulations, appropriate r(C-C) and r(C -C ) restraints were derived, as listed in Tables II and III, and these, rather than the original r(O-O) restraints, are the ones used. The recent [19] NMR study gives complementary information and so was not used here (Loewen et al., 2001). Figure 5.2, Chemical structure of the Cys-nitroxide constructed to determine r(Cα-Cα) restraints from simulations containing r(O-O) restraints between pairs of nitroxide groups Restraint simulations to generate inactive and active conformations The initial receptor model was heated up to 310 K over 50 ps. The initial 2-AR model was then subjected to restrained MD simulations at 310 K in the presence of either inactive or active restraints (Table II and III). In the case of the rhodopsin models, only inactive restraints were applied to the cisretinal bound rhodopsin and only active restraints were applied to the transretinal bound rhodopsin. The restraints were introduced progressively over 100 ps, starting at 1/200 of the restraint energy term and rising progressively every 10 ps. The final 100 ps of MD were conducted in the presence of the restraints at full strength. The structure was then subjected to a conjugate gradient minimisation to generate the final inactive or active receptor model. The stability of these final active and inactive structures were assessed (satisfactorily) by performing 100 ps unrestrained MD simulations at 310 K. Control simulations were run for 1 ns but these structures did not sample any significantly new conformations (as judged by the RMS values). A simulated annealing protocol was also used in which the structures were heated to 500 K. The restraints were introduced progressively as above, with the final 100 ps of MD conducted in the presence of full strength restraints. Structures were collected at 10 ps intervals over the subsequent 100 ps. Each of the 10 structures was then cooled to 310 K over 100 ps. After cooling, the structures were minimized and averaged to one structure that was again minimised to generate a final inactive or active receptor model. This essentially gave no new structures, again indicating that the main simulations were run for sufficient time. Molecular Graphics and structure validation The mutations, loop addition, rotamer analysis and structure validation were carried out using the appropriate modules of WHATIF [56]. In addition, the structures were also validated using the ERRAT program (www.doembi.ucla.edu/Services/ERRAT/), which compares the atom positions in the given structure with identical atoms types in a database derived from x-ray structures, and gives a final residue confidence score. ERRAT highlighted a small number of residues in the models as irregular - but fewer than in the rhodopsin crystal structure. It is common to see residues in X-ray crystal structures highlighted as irregular using this type of analysis. In all other respects, including visual checking to ensure that all potential hydrogen bonding groups were satisfied [32] the structures satisfied the conditions expected from real protein structures (results not shown). Simulations, restraints and parameter derivation The MD simulations were carried out using the Amber united-atom force field as implemented in AMBER5 and AMBER 7(Weiner et al., 1984) using a non- bonded cut off of 12 Å and time step of 0.0005 ps, as in earlier work [36,57,58]. The biochemically derived distance restraints were invoked as either harmonic or half-harmonic restraints, depending on their origin (see Tables II and III), with a force constant of 30 kcal mol-1 Å-2. Half-harmonic distance restraints between backbone carbonyl (n) and amide nitrogen (n+4) of 2.7-3.5Å were applied to ensure that the α helical TM structures was maintained; this allows the helix to sample both α helical and 310 helical conformations. The electrostatic potential derived charges required for retinol and the nitroxide-modified cysteine spin labels were determined using an inhouse version of rattler[59], since this gives charges compatible with AMBER [60]; the additional parameters required were determined by analogy with similar parameters in the AMBER force field. Docking methodology In order to assess the quality of the agonist and antagonist binding sites, we have challenged the models with a set of 172 GPCR ligands using the ligandfit software to determine whether the active model does indeed show a good preference for agonists while the inactive model retains a preference for antagonists. The ligand set included 16 -adrenergic agonists, 38 adrenergic antagonists and 106 other non-peptide opioid, neurokinin, neurotensin and muscarinic ligands primarily obtained from the Cambridge Crystallographic database. Being GPCR ligands, these ligands automatically have ‘drug-like’ properties, and so the only filtering on the ligand selection was to ensure that the adrenergic and non-adrenergic ligands had a similar molecular mass distribution. The ligand binding site was defined using the ligandfit flood filling algorithm [61], with care taken to ensure that the binding site encompassed the area believed to be occupied by all common ligands (as understood from the site-directed mutagenesis data[36]) and that it was sufficiently large to allow the software to select plausible alternative docking modes. The binding site was primed for docking by interactively docking norepinephrine or S-propranolol (the biologically active enantiomer) into the active or inactive models respectively, minimizing, running molecular dynamics for 100 ps and then carrying out a final minimisation. The binding site was also used unprimed and primed with R-propranolol. Given that current virtual screening methods use a rigid enzyme, this step was desirable to ensure that key functional groups such as the serine OH groups on helix 5 etc were orientated to make productive interactions. The docking used softened non-bonded potentials and again this concession was used to make allowances for the standard protocol of using a rigid enzyme, but in other respects the CFF 1.02 force field used by ligandfit is not too dissimilar to that used for the simulations, except that ligand flexibility was implemented by selecting 50000 random trial conformations, ligand polar hydrogens could rotate in increments of 10 and the distance dependent dielectric constant was set to 1/(4r) rather than 1/r. The final ligand conformation was fully minimized within the rigid enzyme. The top ten conformers were saved subject to a restriction that preserves a degree of diversity in the save list. The docked poses were ranked using both the docking score (dockscore; based on van der Waals, electrostatic and ligand internal energy) and the ligscore potentials [60] (ligand fit contains two ligscore potentials, ligscore 1 and ligscore 2; both are based on van der Waals, and surface area terms). Table 5.2 . Relative residue and atomic restraints associated with inactive GPCRs. Experimental evidence rhodopsin X-ray Restraint in receptor model distances Disulfide formation between Cys140(gn345) r(S-S) = 25.5 Å normal r(S-S) restraint or equivalent r(C-C) and Cys316(gn743) in rhodopsin upon restraint between Thr 136 332 (gn345) & Phe (gn743) 25 photo bleaching . Increased rate of disulfide reaction r(S-S) = 7.1 Å between Cys140(gn345) & Cys222(gn527) in normal r(S-S) restraint or equivalent r(C-C) restraint between Thr136(gn345) & Val218(gn527) 84 rhodopsin on photo bleaching Cross-link of retinal ionone ring position 169 r(C-C) = 15.1 Å 3.0 Å < r(C-C) < 4.0 Å; not applicable to 2-AR 28 C-3 to Ala in active state of rhodopsin . Zinc binding to D113C(gn322)/N312H(gn719) r(S-N) = 11 Å 16 r(C-C < 10 Å, r(N-N) < 6 Å; restraint between Asp113(gn322) & Asn312(gn719) mutant 2-AR causes some receptor activity . Increased average distance between r(C-C Å Initial r(O-O) 23 Å restraint between nitroxide V139C(gn344)-K248C(gn601), Å Å reacted with I135C (gn344)-H269C?(gn601), V139C(gn344)-T251C(gn604) and respectyively; I135C (gn344) L272C?(gn604) & I135C (gn344)- V139C(gn344)-R252C(gn605); rhodopsin mutants reacted with nitroxide upon photo K273C?(gn605); subsequent rhodopsin restraints/ r(CC Å Å respectively: Å Å 16.0 > r(C-Cr(CC respectively; 16.0 > r(C-Cr(CC 7 bleaching . 19.6 > r(C-Cr(CC subsequent 2-AR restraints/ Å respectively: 16.2 > r(C-Cr(CC 19.3 > r(C-Cr(CC 15.8 > r(C-Cr(CC No change in distance between r(C-C Å Initially r(O-O) 15-20 Å; restraint between nitroxide V139C(gn344)-E249C(gn602) rhodopsin r(CC Å reacted with I135C (gn344)-K270C?(gn602); mutant reacted with nitroxide upon photo subsequent rhodopsin restraints/ Å respectively: 7 bleaching . 13.3 > r(C-Cr(CC Decrease in V139C(gn344)- r(C-C Initially r(O-O) 12-14 Å; restraint between V250C(gn603) distance; rhodopsin mutant r(CC nitroxide reacted with I135C(gn344)- reacted with nitroxide upon photo A271C?(gn603); bleaching 7. subsequent rhodopsin restraints/ Å respectively: 15.0 > r(C-Cr(CC Expected distance between Asp113(gn322) r(C- C) = 17.0 Å 8 Å < r(C- C) < 10 Å restraint between and Ser204(gn513) derived from ligand Asp113(gn322) & Ser204(gn513) structures 160 . Expected distance between Asp113(gn322) r(C- C) = 15.7 Å 8 Å < r(C- C) < 10 Å restraint between and Ser207(gn516) derived from ligand Asp113(gn322) & Ser207(gn516) structures 160 . Cys285 accessible to aqueous probes upon No applicable Cys285(gn617) accessible to polar pocket. 23 CAM . distance restraint Experimental evidence Rhodopsin X-ray structure distances Restraint in receptor model Disulfide formation between Cys140(gn345) r(S-S) = 7.6 Å normal r(S-S) restraint or 222 & Cys 25 A (gn527) in dark state rhodopsin . equivalent r(C-C) restraint between Thr136(gn345) & Val218(gn527) - but following used insteadA Disulfide formation between H65C(gn140) r(S-S) = 7.1 Å & Cys316(gn743) in dark state rhodopsin 10. normal r(S-S) restraint or equivalent r(C-C) restraint between Phe61(gn140) & Phe336(gn743) Cross-link of retinal 265 C-3 to Trp -ionone ring position r(C-X) 3.8 Å Not applicableC r(CC = 6.5 Å / r(N-N) = 6.4 Å r(C -C 28 in dark state of rhodopsin . Zinc binding site inactivates (gn343) & E268H(gn600) 15 2-AR A134H . ) < 10 Å, N-N < 6 Å; restraint between A134H(gn343) & E268H(gn600). Zinc binding site inactivates 2-AR A134H r(CC = 8.4 Å / r(N-N) = 5.9 Å (gn343) His269(gn601) 15. r(C-C < 10 Å, N-N < 6 Å; restraint between A134H(gn343) His269(gn601) 15 Engineered zinc binding site inactivates 2-AR r(CC = 11.3Å / r(N-N) = 7.2Å A134H(gn343)-L272H(gn604) 15. . r(C-C < 10 Å, N-N < 6 Å; restraint between A134H(gn343)L272H(gn604) D Average distance between V139C(gn344)- r(O(139) - O(248)) = 14.3 Å r(O-O)12-14 Å; restraint K248C(gn601) & V139C(gn344)- r(O(139) - O(251)) = 14.5 Å between nitroxide reacted 135 T251C(gn604); mutants reacted with with Ile 7 269 nitroxide in dark state . His 135 Ile Leu (gn344)- (gn601)D & (gn344)- 272 (gn604) C,D Average distance between V139C(gn344)- r(O(139) - O(249)) = 21.3 Å r(O-O)15-20 Å; restraint E249C(gn602), V139C(gn344)- r(O(139) - O(250)) = 19.3 Å between nitroxide reacted V250C(gn603) & V139C(gn344)- r(O(139) - O(242)) = 19.7 Å with Ile 135 (gn344)- 249 (gn602)D, R252C(gn605); mutants reacted with Lys nitroxide in dark state 7. Ile135(gn344)- Ala250(gn603)D & Ile135(gn344)Lys252(gn605)C,D Engineered zinc binding site in M1 receptor r(CC = 6.3 Å / r(N-N) = 2.6 Å r(C-C < 10 Å, r(N-N) < 6 between L116H(gn333)-S120H(gn337)- r(CC = 10.1 Å / r(N-N) = 4.2 Å Å; restraint between F374H(gn614) 14. r(CC = 13.5 Å / r(N-N) = 7.4 Å Leu124(gn333), Ala128(gn337) 282 & Phe (gn614) E,F Engineered zinc binding site in M1 receptor r(CC = 11.5 Å / r(N-N) = 4.1 Å r(C-C < 10 Å, N-N < 6 Å; between L116H(gn333)-F374H(gn614)- r(CC = 7 Å / r(N-N) = 1.3 Å restraint between N414(gn729) 14. r(CC = 10 Å / r(N-N) = 4.8 Å Leu 124 (gn333), Phe282(gn614) & Asn322(gn729); gn333-gn614 only used in rhodopsin gn333-gn729 not applicable due to bridging water? Engineered zinc binding site in M1 receptor r(CC = 7 Å / r(N-N) = 1.3 Å r(C -C < 10Å, N-N < 6 between F374H(gn614)-N414H(gn729)- r(CC = 6.2 Å / r)N-N) = 2.2 Å Å; restraint between Y418(gn733) 14. r(CC = 11.3 Å / r(N-N) = 1.5 Å Phe282gn614, Asn322(gn729) & Tyr Natural zinc binding site in NK1 receptor 197 between His 265 (gn509)-His (gn622) r(CC = 9.6 Å / r(N-N) = 4.7 Å 158 326 (gn733) E r(C-C < 10Å, N-N < 6 Å; restraint between 200 Ala Increased zinc binding affinity with r(CC = 9.6 Å / r(N-N) = 5.2 Å 290 (gn509)-Phe r(C-C < 10Å, N-N < 6 Å; engineered binding site in NK1 receptor restraint between between E193H(gn505) -Y272H(gn629) Asn 158 196 297 . (gn622) Val (gn505) & (gn629) Increased zinc binding affinity with r(CC = 9.7 Å / r(N-N) = 4.7 Å r(C-C < 10Å, N-N < 6 Å; engineered zinc binding site in NK1 r(CC = 6.1 Å / (N-N) = 3.6 Å restraint between receptor with mutant T201H(gn513) 158. r(CC = 7.8 Å / N-N = 1.1 Å Ala200(gn509)-Phe290(gn622) & Ser204(gn513)E Increased zinc binding affinity with r(CC = 9.7 Å / r(N-N) = 4.7 Å r(C-C < 10Å, N-N < 6 Å; engineered zinc binding site in NK1 r(CC = 5.9 Å / r(N-N) = 2.2 Å restraint between r(CC = 7.3 Å / r(N-N) = 4.8 Å Ala200(gn509)-Phe290(gn622) receptor with mutant L269H(gn626) 158 . & Ile294(gn626)E Interaction of Glu 122 (gn318) & r(CC = 10.3 Å His211(gn516) in rhodopsin 159. r(C-C < 8 Å; restraint between Thr118gn(327) & Ser207(gn516)C 285 Cys inaccessible to aqueous probes 23 B . No applicable distance restraint Cys285(gn617) at helix-helix or helix-lipid interface. A rate of disulfide reaction very slow in the dark state bovine rhodopsin implying a large separation. B the restraint associated with the SCAM data studies on constitutively active mutant 2-AR cannot be included explicitly as a restraint, but served as a check on the final structure. C distance restraints used as a check on the final structure. D Only used in 2-AR because of ligand position. E Restraints not used within the same helix. F only used in rhodopsin. Table 5.3. Relative residue and atomic restraints associated with active GPCRs. Experimental evidence rhodopsin X-ray Restraint in receptor model distances Disulfide formation between Cys140(gn345) r(S-S) = 25.5 Å normal r(S-S) restraint or equivalent r(C-C) restraint and Cys316(gn743) in rhodopsin upon between Thr136(gn345) & Phe332(gn743) 25 photo bleaching . Increased rate of disulfide reaction 140 between Cys 222 (gn345) & Cys r(S-S) = 7.1 Å normal r(S-S) restraint or equivalent r(C-C) restraint between Thr136(gn345) & Val 218(gn527) (gn527) in 84 rhodopsin on photo bleaching Cross-link of retinal -ionone ring position 169 r(C-C) = 15.1 Å 3.0 Å < r(C-C) < 4.0 Å; not applicable to 2-AR r(S-N) = 11 Å r(C-C < 10 Å, r(N-N) < 6 Å; restraint between 28 C-3 to Ala in active state of rhodopsin . Zinc binding to Asp113(gn322) & Asn312(gn719) D113C(gn322)/N312H(gn719) mutant216 AR causes some receptor activity . Increased average distance between r(C-C Å Initial r(O-O) 23 Å restraint between nitroxide reacted V139C(gn344)-K248C(gn601), Å Å with I135C (gn344)-H269C?(gn601), I135C (gn344) V139C(gn344)-T251C(gn604) and respectyively; L272C?(gn604) & I135C (gn344)-K273C?(gn605); V139C(gn344)-R252C(gn605); rhodopsin subsequent rhodopsin restraints/ Å respectively: mutants reacted with nitroxide upon photo r(CC Å 16.0 > r(C-Cr(CC bleaching 7. Å Å 16.0 > r(C-Cr(CC respectively; 19.6 > r(C-Cr(CC subsequent 2-AR restraints/ Å respectively: 16.2 > r(C-Cr(CC 19.3 > r(C-Cr(CC 15.8 > r(C-Cr(CC No change in distance between r(C-C Å Initially r(O-O) 15-20 Å; restraint between nitroxide V139C(gn344)-E249C(gn602) rhodopsin r(CC Å reacted with I135C (gn344)-K270C?(gn602); mutant reacted with nitroxide upon photo subsequent rhodopsin restraints/ Å respectively: 7 bleaching . 13.3 > r(C-Cr(CC Decrease in V139C(gn344)- r(C-C Initially r(O-O) 12-14 Å; restraint between nitroxide V250C(gn603) distance; rhodopsin mutant r(CC reacted with I135C(gn344)- A271C?(gn603); reacted with nitroxide upon photo subsequent rhodopsin restraints/ Å respectively: bleaching 7. 15.0 > r(C-Cr(CC 113 Expected distance between Asp and Ser 204 structures (gn513) derived from ligand & Ser204(gn513) . 113 and Ser 8 Å < r(C- C) < 10 Å restraint between Asp113(gn322) 160 Expected distance between Asp 207 (gn322) r(C- C) = 17.0 Å (gn322) r(C- C) = 15.7 Å (gn516) derived from ligand 8 Å < r(C- C) < 10 Å restraint between Asp113(gn322) structures 285 Cys 160 & Ser207(gn516) . accessible to aqueous probes upon No applicable 23 CAM . Cys285(gn617) accessible to polar pocket. distance restraint 5.3 RESULTS 5.3.1 Inactive receptor conformations Comparison with the X-ray structure. The RMS difference between the energy minimised inactive (cis-retinal bound) rhodopsin model and the crystal structure was 1.6 Å, demonstrating that the structure generated with the restrained MD was very close to the crystal structure. Indeed, the helix tilt angles for both inactive receptor models are very similar to those observed in the crystal structure. Predictably, the greatest differences were observed in the intracellular loops (which are less well-defined in the X-ray structure), primarily intracellular loop 2 (IL2), IL3 and the final 5 residues of helix 8. Nevertheless, the loop structure was stable and the residues Arg175-Thr177 and Cys184-Ala186 (2-AR -sheet formed by numbers) remained throughout both the rhodopsin and 2-AR simulations. The inactive 2-AR receptor model exhibited slightly larger differences from the crystal structure than did the rhodopsin model, with an RMS difference between the backbone atoms of 2.2 Å. The most notable change was in TM7, with the elimination of a stretch of 310 helix around residue gn723 (gn refers to the GPCRDB40 generic numbering scheme defined at www.gpcr.org and in Table I where the first number refers to the helix). This region of the receptor converted into a regular alpha helical structure, with a concomitant anticlockwise rotation (as viewed from the extracellular side of the membrane) of the intracellular part of the helix. The most likely reason for requiring a 310 helix in rhodopsin is the stereochemical restraint62 imposed by the Schiff's base formation between cis-retinal and Lys296; this requirement for a 310 helix does not exist in the 2-adrenerigc receptor. The lack of explicit lipid did not cause the helical bundle to become more compact than the template rhodopsin structure, probably because the interior of the helix bundle and the intraceullular and extracellular loops were solvated. The interhelical distances and tilt angles were comparable to those observed in the rhodopsin crystal structure (results not shown) and other comparable membrane proteins63 . The amphipathic helix (Asp331-Leu342, denoted helix 8, 2-AR numbers) was stable throughout the simulations. Inclusion of nitroxide and histidine zinc binding restraints. The distance between the nitroxide radicals placed at the positions listed in Table II in the initial 2-AR/nitroxide model are similar to those reported in the SDSL studies (Table II). Analysis of the MD trajectories has shown that the nitroxide-reacted cysteine at position Ile135(gn344) is much restricted by the proximity of the intracellular regions of TM5 and TM6; similar observations were made for the five corresponding positions in TM6 (gn601-605). This observation (based on simulations in which only one nitroxide pair was introduced so as to reduce the artefacts of the Cys-nitroxide mutant on the natural protein structure and to mimic the SDSL studies) enabled the spin labelling data to be introduced as C -C restraints. The time averaged nature of the SDSL constraints reduces slightly their utility in defining restraints for use in MD simulations compared to the SDCL studies where a structurally well-defined disulphide bond is formed. However, the results demonstrate that the rhodopsin crystal structure is consistent with the positions of the Cys-nitroxides, since all the distances observed in the model are similar to those obtained experimentally. The engineered histidine zinc binding studies in most cases also agreed well with the crystal structure. As with the restraints derived from the SDSL studies, there is some potential ambiguity associated with these restraints because His and Cys can coordinate zinc through bridging water molecules and as such the restraint lacks absolute definition. Figure 5.5. Overall structural changes on activation for (A) rhodopsin and (B) 2-AR. The bold black and grey lines joins the extracellular helix ends in the inactive and active receptor respectively; the dotted black and grey lines join the corresponding intracellular positions. The transmembrane helices are labelled, clarity permitting, at the mid point between the inactive and active helix ends, in large font for the extracellular ends and small font for the intraceullular ends. The figure primarily shows the increase in the intracellular area on activation. 5.3.2 Active receptor conformations Overall changes. The backbone RMS difference between the final energy minimised active (trans-retinal bound) rhodopsin model and the crystal structure was 6.4 Å; the backbone RMS difference between the final energy minimised active 2-AR receptor model and the rhodopsin crystal structure and the inactive 2-AR model was 7.6 Å and 6.2 Å respectively. The most notable changes on activation were in TM4, TM5, TM6, TM7 and particularly in helix 8, as shown in Fig. 2 and 3; the overall changes are shown schematically in Fig. 4. However, detailed analysis of the structural changes on activation, i.e. the nature of helix translation and rotation was not straightforward as the direction of translation or rotation depends on how the structures were superimposed. (All rotations are described as viewed from the extracellular side of the receptor about an axis roughly perpendicular to the membrane.) Thus, if all the C atoms of the transmembrane helical regions of the active and inactive receptor were superimposed then essentially no rotation was observed for TM5. However, if only TM5 and TM6 were superimposed then there is anticlockwise rotation of TM5 and clockwise rotation of TM6. Alternative superpositions can reverse the apparent rotation, e.g. on TM6. The origin of these apparent anomalies lies partly in the differential rotation for the intracellular end of TM7 and the extracellular end of TM4, partly in the differential proline positions in rhodopsin and 2-AR and partly from the resultant secondary differences in structure. This ambiguity arises also because the true movement is a complex mixture, primarily of translation, but also of bending (especially at proline residues), twisting (e.g. either side of a proline residue) and to a lesser extent rotation. Consequently, while TM6 and TM7 show notable changes on activation, the rotation between TM6 and TM7 is much less than that between TM5 and TM6. These superposition-dependent observations may lead to apparent contradictions between different structural studies that in other respects are in accord with each other. Nevertheless, several generalizations can be made. TM4 shows a large clockwise rotation in the rhodopsin model because of the cross-link from retinal to Ala169 in active state of rhodopsin28. Since there is no corresponding photoaffinity labelling data for the 2-AR, this restraint was only applied to rhodopsin. Nevertheless, the other restraint data results in a diminished but still significant clockwise rotation of TM4. TM4 also moves slightly away from the bundle but this is probably because there is less restraint data available for TM4 than for other helices. TM5 shows little evidence of rotation, except with respect to TM6; inactive TM5 is essentially straight, but bends on activation at the highly conserved 2-AR Pro211 (gn520) (97% in the amine family). Initially, a kink around 2-AR Pro288(gn620), which is part of the CWXP motif and 99% conserved in the amine family , allows either end of TM6 to interact with opposite faces of TM3. On activation, the intracellular end of TM6 moves away from TM2 and TM3 as the Arg(gn340)-Glu(gn600) salt-bridge is broken and in the process, TM6 straightens out (Mutation of Pro in TM6 causes constitutive act in the alpha factor receptor64). The straightening of TM6 allows TM5 to move closer and to some extent the bending in TM5 in the active state mirrors that in TM6. The consequential reduction in the Cys265 (TM6) – Lys 224 (TM5) distance on activation, which Ghanouni related to an increase in fluorescence quenching in 2-AR, can therefore be interpreted primarily as a change in the tilt angle of helix 6 rather than as a clockwise rotation65. Similarly, Cys 265 is accessible in the active but not the inactive structure and so the SCAM results on a constitutively active 2-AR mutant can also be explained by a change in the tilt angle rather than an anticlockwise rotation of TM666. The consequential increase in solvent exposure on activation between the intracellular parts of helices 3 and 6 has been noted elsewhere67-70. The rotation, translation and bending of TM6 permits key residues to have differential accessibility towards agonists and antagonists in the active and inactive state respectively, as discussed below. Helix 7 is essentially straight in the inactive state36, the most recent cryoelectron microscopy structure71,72 and the X-ray structure1 but the simulations show that it kinks on activation as the intracellular end moves towards TM3 (as TM6 moves away) to help form the Cys 140-Cys316 rhodopsin disulphide restraint. The flexibility of this region is perhaps due to the Asn322Pro323(gn730) motif which lies in the vicinity of the kink originally described by Fu et al19,34 (Fu and Konvicka’s controversial kink proposal 36 originally applied to the inactive dopamine D2 receptor (which in certain expression systems has a high level of basal activity, J. A. Javitch, personal communication)) and is a classic case of how discrepancies between models can be resolved once the data is rigorously separated into that relevant to active models and that relevant to inactive models). Indeed, artificially introducing this kink enables the receptor to more readily satisfy the active restraints (results not shown). This movement of TM7 is the only movement that reduces the exposed intracellular surface area. However, since the Cys140-Cys316 rhodopsin disulphide bond is formed under artificial conditions (i.e. on addition of oxidizing agent) it seems that while this cross-link can form, it does not necessarily form in vivo. The consequent large movement seen in Helix 8, driven by the restraint between Cys 140(gn345) and Cys316(gn743) aided by the kink in TM7, may therefore be interpreted as an increase in mobility of this region. TM2 shows comparatively less movement, but the intracellular end still moves out on activation from a very central position under the helical bundle close to the intracellular ends of all helices apart from TM5; this movement is aided by a reduction in the bend at Pro88(gn233) so that the intracellular end moves away from TM6. (Pro88 is 85% conserved in the biogenic amine receptors but not in the opsins, some of which have a weakly conserved (~30%) GG motif at a similar position (gn230, 231)). Interactions of Trp286 (gn618). The conserved Trp286 (gn618, 97% conserved in the Amines) on TM6 interacts plays a key role as it interacts with TM3 and TM7 in the inactive model but moves away from TM7 to interact with TM5 in the active model. (It also interacts with the ligand in many systems.) A glycine in TM7, Gly315 (gn722, 75% conserved in Amines), provides an ideal space for the indole ring of Trp286 (gn618) to occupy, thus reducing steric clashes. Interestingly, position gn722 is 100% conserved as Gly in the adrenergic and dopamine receptors, 100% by Ala in the tachykinin receptors, 97% by Ala in rhodopsin but 100% by Phe in the neurotensin receptors. Mutation of this Phe to Ala in the human neurotensin NT2 receptor reduces the constitutive activity associated with this subtype (P. Angeloz-Nicoud, personal communication). This implies that a small amino acid (Ala, Gly, Ser, Cys) is required at this position to correctly place Trp(gn618) in the inactive state of the receptor. Gly121 and Phe261 forma a similar pair in the opsins, as shown by compensating mutations in which increases in size at the glycine are compensated by a decrease in size at the phenylalanine73,74. Table 5.4. Hit rates, yields and enrichment factors for adrenergic agonists and antagonists, along with the maximum possible values that would be obtained with perfect structures and software. The enrichment factor is the number of ligands determined divided by the number expected for a random process (indicated by the dashed line in fig. 5). Hit Yield Hit rate Enrichment list (%) (%) Factor (%) Obs. Max Obs Max Obs Max Agonists (max 16): active model 5 38 50 75 100 5.7 7.7 10 50 100 50 100 4.4 8.7 15 63 100 42 100 4.0 6.3 20 88 100 44 100 4.1 4.7 25 88 100 35 100 3.4 3.9 Ave 65 90 49 100 4.3 6.3 Antagonist (max 38) S-propranolol-based model 5 11 21 50 100 2.1 4.2 10 21 42 50 100 2.1 4.2 15 32 63 50 100 2.1 4.2 20 37 84 44 100 1.8 4.2 25 39 100 38 100 1.6 4.0 Ave 28 62 46 100 1.9 4.2 Antagonist (max 28) apoprotein-based model 5 4 32 11 100 0.4 4.0 10 18 57 31 100 1.3 4.1 15 36 86 42 100 1.9 4.5 20 50 100 45 100 2.1 4.1 25 54 100 39 100 1.8 3.3 Ave 32 75 34 100 1.5 4.0 Antagonist (max 38): R-propranolol-based model 5 8 21 38 100 1.6 4.3 10 16 42 38 100 1.6 4.3 15 18 63 29 100 1.3 4.3 20 32 84 38 100 1.6 4.3 25 39 100 38 100 1.6 4.1 Ave 23 62 36 100 1.6 4.3 Agonist (max 16): active model (hybrid) 5 19 50 38 100 2.9 7.7 10 50 100 50 100 4.4 8.7 15 56 100 38 100 3.6 6.3 20 75 100 38 100 3.5 4.7 25 81 100 33 100 3.2 3.9 Ave 56 90 39 100 3.5 6.3 Antagonist (max 28): apo-based model (hybrid) 5 10 15 20 25 Ave 7 14 32 46 50 30 32 57 86 100 100 75 22 25 38 42 37 33 100 100 100 100 100 100 0.9 1.0 1.7 1.9 1.7 1.4 4.0 4.1 4.5 4.1 3.3 4.0 5.3.3 Docking results The overall docking results for the most highly ranked conformations are shown in Fig. 5. In order to compare these results to Bissantz et al [75], we have generated 5 hit lists taken from the top 5% - top 25% most highly ranked ligands and determined the hit rate (the proportion of true hits in the hit list) and the yield (the proportion of true hits in the hit list compared to the full ligand list) with a view to determining the suitability of these structures, particularly the active one, for virtual screening, bearing in mind that while good results do not necessarily by themselves validate the model, they do contribute towards this end. The results are tabulated in Table IV, along with the maximum possible yields, hit rates and enrichment factors that are attainable with the given number of ligands. The antagonist results in Fig. 5A and 5B show that there is little enrichment until about rank 10; the false positives in this region are primarily opioid ligands. Given the known interactions between adrenergic and opioid receptors [76,77], it is possible that these are true hits, especially as both receptors share an Asp at position gn322, but we are treating the highly ranked opioid ligands as false positives. Such observations of very highly ranked false positives are quite common in docking studies. For the agonist results in Fig. 5C there is immediate enrichment and the results are very good compared to related studies [75]. Careful examination of the docking mode of the adrenergic agonists and antagonists shows that the results are generally in accord with the known binding mode as determined by site directed mutagenesis and discussed elsewhere [36,75]. We observed that while ligscore gave very poor enrichment factors (with ligscore 2 giving only marginal improvement over ligscore 1 in these systems), ligscore 1 was superior to dockscore at selecting the correct docking mode from a set of alternative docked conformations. Consequently, we have also ranked the structures according to the dockscore for the best ligscore conformation. The hit rates, yields and enrichment factors from this hybrid approach are only slightly inferior to the pure dockscore results but are based on superior structures for some compounds. While slightly better docking results were obtained when the inactive models were minimized in the presence of an appropriate ligand (norepinephrine or propranolol), the results on the apoprotein-based models provided additional evidence that the lack of explicit lipid did not cause the helical bundle to become more compact than the template rhodopsin structure. It is encouraging that slightly higher enrichment factors were obtained when the inactive model was minimized in the presence of Spropranolol (the biologically active form) whereas slightly lower enrichment factors were obtained when the inactive model was minimized in the presence of R-propranolol. 5.4 Discussion 5.4.1 Palmitoylation in the inactive and active receptor models Many GPCRs have palmitoylation sites; in the case of the β2-AR receptor the palmitate is attached to the C-terminal Cys341 residue[78-81] located at the end of the amphipathic helix 8, consistent with the position of helix 8 lying at the membrane surface in both the crystal structures and the inactive models[78,82,83]. Exposure of GPCRs to agonists (or other factors such as NO) generally increases the rate of palmitoylation and this usually results in an increase in palmitoylation levels but can also lead to no change or a decrease, depending on the kinetics [79]. The functional consequences of this turnover are difficult to establish as mutagenesis removes both the cysteine residue and the palmitate. In the active receptor model, the Cys140(gn345) and Cys316(gn743) rhodopsin disulfide restraint drives Cys 341 on helix 8 to interact with the intracellular extremities of TMs 3, 5 and 6, as shown in Fig. 2. Whether these interactions are likely to be formed continuously or intermittently in the active structure is not clear, as Cu (phenantholine)3+ was required as oxidant to form the disulphide bond [84], but the active model structure shown in Fig. 2. may not be consistent with the covalent attachment of palmitate, particularly for receptors palmitoylated at positions N-terminal of Cys341. If the interactions are formed continuously, then helix 8 will lie below the helical bundle with possible negative structural consequences for G-protein coupling as it would partially block access to the exposed region between TM3 and TM6 observed here and by others who have related this opening to G-protein coupling [67-70]. Such a blocked conformation could underlie the reduced G-protein coupling observed in some depalmitoylated receptors79 but the reduced G-protein coupling is probably due to the phosphorylation of depalmitoylated receptors [79]; a second unlikely alternative consequence is that depalmitoylation could be a prerequisite for activation as helix 8 may need to leave the membrane surface but this is not consistent with other observations [79]. While Oprian and coworks did check that some disulphide-linked split rhodopsin receptors activated transducin84, this was not checked for the gn345-gn316 disulfidelinked receptor25. A more likely scenario then is that the gn345-gn316 interactions are intermittent, made possible by the movement of TM6 away from TM3, in which case only the increased mobility of TM7 and helix 8, and the space between TM3 and TM6 required to give access to Cys 316, are characteristic of activation. In this case, palmitoylation may be required to either prevent phosphorylation or keep helix 8 away from intracellular loops 2 and 3 and thus preserve G-protein coupling. We note here that the possible palmitoylation [79,85] site on intracellular loop 2 is accessible in both active and inactive forms of the receptor. 5.4.2 Preferential interactions with agonists and antagonists Residue positions gn621 and gn622 are important as mutation of either can affect ligand binding and signalling [86,87]. Site directed mutagenesis data indicates that in many receptor families, residue position gn621 is associated with agonist binding and signal transduction [64,88-99] (in that these effects are generally reduced or abolished in mutant receptors) while antagonist binding is not affected by gn621 mutation [100]. Position gn622 is generally associated with antagonist binding [101-104] and mutation generally does not affect agonist binding [102,105-109]. In a small number of studies both residues have been mutated and contrary effects have been observed, e.g. mutation of gn622 affecting agonist binding [106,110-112] or does not affect antagonist binding 113 and mutation of gn621 affecting antagonists but not agonists [114] (in some cases the effect of mutation on agonists at gn622 is less than that at gn62198). While, it has also been observed that mutation of gn621 can affect antagonists [115], these are usually non-peptide antagonists of peptide receptors [115,116]. In some comprehensive studies, mutation of gn621 only affects agonists while mutation of gn622 only affects antagonists [117,118] but in other studies the situation is not as clear-cut, partially because different agonists and antagonists have different effects. SCAM studies are equivocal – one indicates that both gn621 and gn622 are accessible to the binding pocket [119] while another indicates that only gn621 is accessible88; the increased exposure of gn621 is in agreement with this overall picture. Similar associations of gn718 with agonist binding [36,120,123] and gn719 with antagonist binding [36,96,124,132] have also been made, but again the situation is not clear cut and only a few comprehensive studies have been made, partly because mutation of gn719 can lead to misfolded [127] or poorly expressed receptors [104]. Exceptions include indications that both residues are involved in antagonist binding [87] or that mutation at gn719 affects agonists[112,133-136] or agonist and antagonist binding(but doesn’t necessarily stop signalling) [31,137-141] or binds an inverse agonist in a constitutively active receptor [142] or does not affect antagonist binding [113,134,143]. Some of these anomalous results may arise because some inverse agonists, which may prefer to bind to the active state, have been described as antagonists (the lower enrichment factors for antagonists compared to agonists (see Fig. 5. and Table IV) may also derive from the labelling of inverse agonists as antagonists). Our model structures are partially in agreement with these observations in that while gn621 and gn719 are exposed in both active and inactive structures, gn622 becomes much less exposed in the active structures while gn718 becomes more exposed in the rhodopsin active structure, partly as a result of rotational and translational movement, but also through proline bending in TM6. The relationship between these observations and our structural model is partially obscured by the differing helical structure and flexibility of helix 7 in the two models, arising from the differing 310 helix content which results in different degrees of rotation of TM7. This inherent flexibility of TM7 may be the origin of the less than clear site-directed mutagenesis effects at these important loci, as indicated by site-directed mutagenesis effects on ligand binding also being observed at position gn720 [126,144] . These loci have been implicated as conformational switching regions, partly because certain amino acid mutations at position gn719 transform antagonists into agonists [96,129] or lead to constitutive activation[107,145]. Nevertheless, the magnitude of the observed changes in this region of the receptor are less than those observed at the intracellular end of helices 6 and 7. Residue gn722, which is directly below gn719, is also primarily associated with agonist rather than antagonist binding[146-148], though again there are exceptions where there is either little effect [90,105,144] or a preferential effect on antagonists [114]. Again, the increased exposure of gn722 on activation, like that of gn71, is linked to the movement of gn621 that also helps to reduce the exposure of gn622 (and Trp gn618). Likewise, Cys(gn617) is not exposed to the binding cavity in the inactive receptor becomes exposed on activation, partly as a result of the movement of gn621 and Trp(gn618); this residue has been shown by SCAM methodology to be exposed in a constitutively active mutant [66], but not in the ordinary receptor [119]. 5.4.3 The requirement for multiple GPCR models Pharmacological studies show that agonists, antagonists and inverse agonists do not bind to a single receptor conformation, an observation eloquently elaborated in Gether's recent review [149] of the two state (R/R*) model of GPCR activation [150]. In contrast, earlier models of GPCRs, typically those constructed from cryoelectron microscopy data [151], failed to distinguish rigorously between an inactive and an active conformation [32,33]. In many cases, one model was used for docking all classes of ligand: agonist, antagonist and inverse agonist. A comparison of the available models reveals significant differences, usually in the relative helix rotations, helix length and the depth of key residues within the (implicit) membrane. Some of these differences can be attributed to the fact that data from biophysical studies and site-directed mutagenesis studies on agonist, antagonist and inverse agonist binding and activity are incorporated into a single model. In contrast, pharmacological, biophysical and structural data, and data on constitutively active receptors [152], all demonstrate that GPCRs exist in distinguishable conformations and hence a single model for any receptor can never be adequate. The usual method to generate an active model is to simulate the conformation changes induced in the presence of an agonist or in the presence of mutations that generate constitutive activity. However, because of limitations in phase space sampling, it is unlikely that the resultant changes from such approaches would be transmitted throughout the structure. Because we have used restraints from most regions of the receptor to generate an active conformation, the problems arising from phase space sampling are likely to be less of a problem with this approach. 5.4.4 Constraining interactions Various groups have suggested that the inactive conformation of a receptor is maintained through a network of constraining interactions [153155] and that disruption of one of these key constraining interactions might result in a degree of activity being observed [155]. This hypothesis is reinforced by the observation that mutations that cause constitutive activity for a unique receptor signalling pathway can be observed in receptors that naturally couple to multiple distinct signalling pathways [156]. Our simulations support the idea of constraining interactions and here the salt bridge between Arg(gn340) and Glu(gn600) presented itself as the main constraining interaction. (Both of these residues are present in the 1F88 crystal structure in a sparsely populated rotameric form, but can only interact directly in the refined structure [157] by rotation about torsion 1 of the rhodopsin Glu247(gn600) side chain). In certain "active" simulations, this salt bridge did not break and the final structure did not readily satisfy the active restraints. On the other hand, simulations in which this interaction was broken by an E268A ( 2-AR) mutation went a long way towards satisfying the active restraints even if they were not applied (results not shown). Likewise, Arg (gn330) – Glu (gn600) was the only restraining interaction that had to be artificially broken in order to satisfy the active restraints. Constraining interactions, if not dealt with adequately, e.g. through active constraints applied throughout the structure, present a reason why an active receptor model may retain inactive character. A related observation is the fact that TM6 is alone in having only a single hydrogen bond to other helices [157], ensuring that there are few constraining interactions. This supports the dynamic role of TM6 in the activation mechanism. 5.4.5 Docking Generally the docking results presented in Table IV compare very favourably with those of Biassantz et al., who showed positive evidence that GPCR models can be used in virtual screening [75]. Biassantz et al. generally obtained lower hit rates 5-39% and yields (0-70%), particularly for agonists. The following results are particularly relevant. Here we have used a single docking method rather than an elegantly combined consensus approach, which would tend to increase the relative quality of Bissantz’ results. In addition, our ligand list is comprised entirely of GPCR ligands, and has some ligands that are very similar to the -adrenergic ligands, including - adrenergic and opioid ligands, which both bind to a highly conserved aspartate on helix 3: these factors would tend to bias our results towards lower enrichment, whereas the Biassantz list contained a significantly more diverse set of compounds. The ligandfit approach used here has been shown to yield results competitive to other methods [61], but we have no evidence to suggest it is vastly superior to other methods and so the very favourable results produced here can be partially attributed to the method used to generate the active and inactive structures. It is particularly satisfying that our active structure generates such good results as this represents further divergence away from its rhodopsin parent structure than for the inactive structure. The lower enrichment for the inactive structure is partly a consequence of our observation that the agonists tend to bind to both the active and inactive structure while the antagonists tend not to bind to the active structure (and this is not merely related to size). These docking control studies therefore produce good evidence to support both the structures produced, and the method used to generate them. 5.5 CONCLUSIONS Sets of experimental distance restraints were obtained from published site-directed cross-linking, engineered zinc binding, site-directed spinlabelling, IR spectroscopy and photo-affinity labelling experiments and rigorously separated into restraints relevant to inactive receptors and those relevant to active receptors. Preliminary simulations showed that the nitroxide labelled substituted cysteine side chains had limited mobility, permitting incorporation of this data as C -C restraints. Molecular dynamics simulations in the presence of either “active” restraints or “inactive” restraints were used to generate two distinguishable receptor models. The active model was satisfactorily generated only after giving due care to releasing the restraining interaction between Arg (gn330) – Glu (gn600). The main changes in the receptor conformation on activation involve TM4, TM5, TM6 and TM7. It is esoterically disappointing that rhodopsin does not show exactly the same changes on activation as the 2-adrenergic receptor (see Fig. 4), but the main origins of this lie in the different 310-helix content on TM7 and the photoaffinity labelling-derived restraints on TM4 used only for rhodopsin. Nevertheless, the main changes are similar and included significant clockwise rotation of TM4, the displacement of TM6 away from TM3 accomplished by a straightening of TM6 so that it interacts more closely with TM5, increased flexibility in the intracellular half of TM7 and a general opening of the intracellular part of the structure. The differences in structure on activation have been related to selected key loci, namely positions gn621/gn622 and gn718/gn719 that play different roles in the active and inactive structures. The 2-AR active model, in particular, gave high yields, hit rates and enrichment factors for selecting agonists from related GPCR drug-like ligands in a virtual screen using the ligandfit docking software. These docking results help to validate both the models and the process used to derive them. Our overall conclusions from the docking experiments are in line with those of Bissantz [75], namely that homology models of G-protein coupled receptors offer potential in the rational design of ligands. However, there are still regions such as TM4 where the determination of additional active restraints could significantly improve the modeling process. In the past, many discrepancies have arisen between models and between models and experiment as a result of failing to identify clearly whether supporting experimental data applies to the active or inactive state; a good example of this is the proposed kink[19,34,36] at the NPXXY motif in TM7 which we propose here applies only to the active state. Acknowledgments It should be noted that the author of this thesis did not carry out the construction of the two adrenergic receptor models (these were constructed by N. J. Kidley and P. Gouldson). The same models have been used for the worked described in Chapter 6 as an template structure under the simulation of the membrane environment with Charmm and a similar methodology was used for the construction of the .The work in this chapter has been published in Proteins Structure, Function & Bioinformatics 2004, Jul 1, 56:67-84 and Bio (GR), 2005, Jan 1, 12: 26-33 Reference List 1. Palczewski, K, Kumasaka, T, Hori, T, Behnke, CA, Motoshima, H, Fox, BA, Le, T, I, Teller, DC, Okada, T, Stenkamp, RE, Yamamoto, M,.Miyano, M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 2000; 289:739-745. 2. Altenbach, C, Yang, K, Farrens, DL, Farahbakhsh, ZT, Khorana, HG,.Hubbell, WL. Structural features and light-dependent changes in the cytoplasmic interhelical e-f loop region of rhodopsin - a site-directed spinlabeling study. Biochemistry 1996; 35:12470-12478. 3. Altenbach, C, Cai, K, Khorana, HG,.Hubbell, WL. Structural features and light-dependent changes in the sequence 306-322 extending from helix VII to the palmitoylation sites in rhodopsin: a site-directed spin-labeling study. Biochemistry 1999; 38:7931-7937. 4. Altenbach, C, Klein-Seetharaman, J, Hwa, J, Khorana, HG,.Hubbell, WL. Structural features and light-dependent changes in the sequence 59-75 connecting helices I and II in rhodopsin: a site-directed spin-labeling study. Biochemistry 1999; 38:7945-7949. 5. Cai, K, Klein-Seetharaman, J, Farrens, D, Zhang, C, Altenbach, C, Hubbell, WL,.Khorana, HG. Single-cysteine substitution mutants at amino acid positions 306-321 in rhodopsin, the sequence between the cytoplasmic end of helix VII and the palmitoylation sites: sulfhydryl reactivity and transducin activation reveal a tertiary structure. Biochemistry 1999; 38:7925-7930. 6. Farahbakhsh, ZT, Ridge, KD, Khorana, HG,.Hubbell, WL. Mapping lightdependent structural changes in the cytoplasmic loop connecting helices C and D in rhodopsin: a site-directed spin labeling study. Biochemistry 1995; 34:8812-8819. 7. Farrens, DL, Altenbach, C, Yang, K, Hubbell, WL,.Khorana, HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. <EM>Science</EM> 1996; 274:768-770. 8. Klein-Seetharaman, J, Hwa, J, Cai, K, Altenbach, C, Hubbell, WL,.Khorana, HG. Single-cysteine substitution mutants at amino acid positions 55-75, the sequence connecting the cytoplasmic ends of helices I and II in rhodopsin: reactivity of the sulfhydryl groups and their derivatives identifies a tertiary structure that changes upon light-activation. Biochemistry 1999; 38:7938-7944. 9. Langen, R, Cai, K, Altenbach, C, Khorana, HG,.Hubbell, WL. Structural features of the C-terminal domain of bovine rhodopsin: a site-directed spin-labeling study. Biochemistry 1999; 38:7918-7924. 10. Yang, K, Farrens, DL, Altenbach, C, Farahbakhsh, ZT, Hubbell, WL,.Khorana, HG. Structure and function in rhodopsin - cysteines 65 and 316 are in proximity in a rhodopsin mutant as indicated by disulfide formation and interactions between attached spin labels. Biochemistry 1996; 35:14040-14046. 11. Yang, K, Farrens, DL, Hubbell, WL,.Khorana, HG. Structure and function in rhodopsin. Single cysteine substitution mutants in the cytoplasmic interhelical E-F loop region show position-specific effects in transducin activation. Biochemistry 1996; 35:12464-12469. 12. Elling, CE,.Schwartz, TW. Connectivity and orientation of the seven helical bundle in the tachykinin NK-1 receptor probed by zinc site engineering. EMBO Journal 1996; 15:6213-6219. 13. Elling, CE, Thirstrup, K, Nielsen, SM, Hjorth, SA,.Schwartz, TW. Engineering of metal-ion sites as distance constraints in structural and functional analysis of 7tm receptors. Folding & Design 1997; 2:S76-S80. 14. Lu, ZL,.Hulme, EC. A network of conserved intramolecular contacts defines the off-state of the transmembrane switch mechanism in a seventransmembrane receptor. J Biol Chem 2000; 275:5682-5686. 15. Sheikh, SP, Vilardarga, JP, Baranski, TJ, Lichtarge, O, Iiri, T, Meng, EC, Nissenson, RA,.Bourne, HR. Similar structures and shared switch mechanisms of the beta2-adrenoceptor and the parathyroid hormone receptor. Zn(II) bridges between helices III and VI block activation. J Biol Chem 1999; 274:17033-17041. 16. Elling, CE, Thirstrup, K, Holst, B,.Schwartz, TW. Conversion of agonist site to metal-ion chelator site in the beta(2)-adrenergic receptor. Proc Natl Acad Sci U S A 1999; 96:12322-12327. 17. Rosenkilde, MM, Lucibello, M, Holst, B,.Schwartz, TW. Natural agonist enhancing bis-His zinc-site in transmembrane segment V of the tachykinin NK3 receptor. FEBS Letters 1998; 439:35-40. 18. Thirstrup, K, Elling, CE, Hjorth, SA,.Schwartz, TW. Construction of a high affinity zinc switch in the kappa-opioid receptor. J Biol Chem 1996; 271:7875-7878. 19. Fu, D, Ballesteros, JA, Weinstein, H, Chen, J,.Javitch, JA. Residues in the seventh membrane-spanning segment of the dopamine D2 receptor accessible in the binding-site crevice. Biochemistry 1996; 35:1127811285. 20. Javitch, JA, Li, X, Kaback, J,.Karlin, A. A cysteine residue in the third membrane-spanning segment of the human D2 dopamine receptor is exposed in the binding- site crevice. Proc Natl Acad Sci U S A 1994; 91:10355-10359. 21. Javitch, JA, Fu, DY,.Chen, JY. Residues in the fifth membrane-spanning segment of the dopamine d2 receptor exposed in the binding-site crevice. Biochemistry 1995; 34:16433-16439. 22. Simpson, MM, Ballesteros, JA, Chiappa, V, Chen, JY, Suehiro, M, Hartman, DS, Godel, T, Snyder, LA, Sakmar, TP,.Javitch, JA. Dopamine D4/D2 receptor selectivity is determined by a divergent aromatic microdomain contained within the second, third, and seventh membranespanning segments. Mol Pharmacol 1999; 56:1116-1126. 23. Javitch, JA, Fu, DY, Liapakis, G,.Chen, JY. Constitutive activation of the beta(2) Adrenergic receptor alters the orientation of its sixth membranespanning segment. J Biol Chem 1997; 272:18546-18549. 24. Kono, M, Yu, H,.Oprian, DD. Disulfide bond exchange in rhodopsin. Biochemistry 1998; 37:1302-1305. 25. Yu, H, Kono, M,.Oprian, DD. State-dependent disulfide cross-linking in rhodopsin. Biochemistry 1999; 38:12028-12032. 26. Zeng, FY, Hopp, A, Soldner, A,.Wess, J. Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. J Biol Chem 1999; 274:16629-16640. 27. Struthers, M, Yu, H, Kono, M,.Oprian, DD. Tertiary interactions between the fifth and sixth transmembrane segments of rhodopsin. Biochemistry 1999; 38:6597-6603. 28. Borhan, B, Souto, ML, Imai, H, Shichida, Y,.Nakanishi, K. Movement of retinal along the visual transduction path. Science 2000; 288:2209-2212. 29. Darrow, JW, Hadac, EM, Miller, LJ,.Sugg, EE. Structurally similar small molecule photoaffinity CCK-A agonists and antagonists as novel tools for directly probing 7TM receptor-ligand interactions. Bioorg Med Chem Letts 1998; 8:3127-3132. 30. Dong, MQ, Dings, XQ, Pinon, DI, Hadac, EM, Oda, RP, Landers, JP,.Miller, LJ. Structurally related peptide agonist, partial agonist, and antagonist occupy a similar binding pocket within the cholecystokinin receptor - Rapid analysis using fluorescent photoaffinity labeling probes and capillary electrophoresis. J Biol Chem 1999; 274:4778-4785. 31. Ji, ZS, Hadac, EM, Henne, RM, Patel, SA, Lybrand, TP,.Miller, LJ. Direct identification of a distinct site of interaction between the carboxyl-terminal residue of cholecystokinin and the type A cholecystokinin receptor using photoaffinity labeling. J Biol Chem 1997; 272:24393-24401. 32. Pogozheva I.D., Lomize A.L.,.Mosberg H.I. The transmembrane 7- bundle of rhodopsin: distance geometry calculations with hydrogen bonding constraints. Biophys J 1997; 72:1963-1985. 33. Scheer, A,.Cotecchia, S. Constitutively active g protein-coupled receptors potential mechanisms of receptor activation. Journal of Receptor & Signal Transduction Research 1997; 17:57-73. 34. Konvicka, K, Guarnieri, F, Ballesteros, JA,.Weinstein, H. A proposed structure for transmembrane segment 7 of G protein-coupled receptors incorporating an asn-Pro/Asp-Pro motif. Biophys J 1998; 75:601-611. 35. Zhou, W, Flanagan, C, Ballesteros, JA, Konvicka, K, Davidson, JS, Weinstein, H, Millar, RP,.Sealfon, SC. A reciprocal mutation supports helix 2 and helix 7 proximity in the gonadotropin-releasing hormone receptor. Mol Pharmacol 1994; 45:165-170. 36. Gouldson, PR, Snell, CR,.Reynolds, CA. A new approach to docking in the beta 2-adrenergic receptor that exploits the domain structure of G-proteincoupled receptors. J Med Chem 1997; 40:3871-3886. 37. Donnelly, D, Findlay, JB,.Blundell, TL. The evolution and structure of aminergic G protein-coupled receptors. Receptors & Channels 1994; 2:6178. 38. Donnelly, D, Maudsley, S, Gent, JP, Moser, RN, Hurrell, CR,.Findlay, JB. Conserved polar residues in the transmembrane domain of the human tachykinin NK2 receptor: functional roles and structural implications. Biochemical Journal 1999; 339:55-61. 39. Horn, F, Weare, J, Beukers, MW, Horsch, S, Bairoch, A, Chen, W, Edvardsen, O, Campagne, F,.Vriend, G. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res 1998; 26:275-279. 40. Oliveira, L, Paiva, AM,.Vriend, G. A common motif in G-protein-coupled seven transmembrane helix receptors. J Comput -Aided Mol Design 1993; 7:649-658. 41. Harris, M, Kihlen, M,.Bywater, RP. PLIM: a protein-ligand interaction modeller. Journal of Molecular Regonition 1993; 6:111-115. 42. Iadanza, M, Holtje, M, Ronsisvalle, G,.Holtje, HD. kappa-opioid receptor model in a phospholipid bilayer: Molecular dynamics simulation. J Med Chem 2002; 45:4838-4846. 43. Tang, P,.Xu, Y. Large-scale molecular dynamics simulations of general anesthetic effects on the ion channel in the fully hydrated membrane: The implication of molecular mechanisms of general anesthesia. Proc Natl Acad Sci U S A 2002; 99:16035-16040. 44. Tajkhorshid, E, Nollert, P, Jensen, MO, Miercke, LJW, O'Connell, J, Stroud, RM,.Schulten, K. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science 2002; 296:525-530. 45. Im, W,.Roux, B. Ions and counterions in a biological channel: A molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J Mol Biol 2002; 319:11771197. 46. Im, W,.Roux, B. Ions and counterions in a biological channel: A molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J Mol Biol 2002; 319:11771197. 47. Capener, CE,.Sansom, MSP. Molecular dynamics simulations of a K channel model: Sensitivity to changes in ions, waters, and membrane environment. Journal of Physical Chemistry B 2002; 106:4543-4551. 48. Rohrig, UF, Guidoni, L,.Rothlisberger, U. Early steps of the intramolecular signal transduction in rhodopsin explored by molecular dynamics simulations. Biochemistry 2002; 41:10799-10809. 49. Kong, YF,.Ma, JP. Dynamic mechanisms of the membrane water channel aquaporin-1 (AQP1). Proc Natl Acad Sci U S A 2001; 98:14345-14349. 50. Dehner, A, Planker, E, Gemmecker, G, Broxterman, QB, Bisson, W, Formaggio, F, Crisma, M, Toniolo, C,.Kessler, H. Solution structure, dimerization, and dynamics of a lipophilic alpha/3(10)-Helical, C-alphamethylated peptide. Implications for folding of membrane proteins. Journal of the American Chemical Society 2001; 123:6678-6686. 51. Goetz, M, Carlotti, C, Bontems, F,.Dufourc, EJ. Evidence for an alphahelix -> pi-bulge helicity modulation for the neu/erbB-2 membranespanning segment. A H-1 NMR and circular dichroism study. Biochemistry 2001; 40:6534-6540. 52. Johnson, ET,.Parson, WW. Electrostatic interactions in an integral membrane protein. Biochemistry 2002; 41:6483-6494. 53. Lin, JH, Baker, NA,.McCammon, JA. Bridging implicit and explicit solvent approaches for membrane electrostatics. Biophysical Journal 2002; 83:1374-1379. 54. Higaki, JN, Fletterick, RJ,.Craik, CS. Engineered metalloregulation in enzymes. Trends in Biochemical Sciences 1992; 17:100-104. 55. Gouldson, PR, Snell, CR,.Reynolds, CA. A new approach to docking in the 2-adrenergic receptor that exploits the domain structure of G-proteincoupled receptors. J Med Chem 1997; 40:3871-3886. 56. Vriend, G. WHATIF: A molecular modelling and drug design program. J Mol Graph 1990; 8:52-56. 57. Gouldson, PR,.Reynolds, CA. Simulations on dimeric peptides - evidence for domain swapping in g-protein-coupled receptors. Biochem Soc Trans 1997; 25:1066-1071. 58. Gouldson, PR, Snell, CR, Bywater, RP, Higgs, C,.Reynolds, CA. Domain swapping in G-protein coupled receptor dimers. Protein Engng 1998; 11:1181-1193. 59. Ferenczy, GG, Reynolds, CA,.Richards, WG. Semiempirical AM1 electrostatic potentials and AM1 electrostatic potential derived charges - a comparison with abinitio values. Journal of Computational Chemistry 1990; 11:159-169. 60. Reynolds, CA, Ferenczy, GG,.Richards, WG. Methods for determining the reliability of semiempirical electrostatic potentials and potential derived charges. Theochem-Journal of Molecular Structure 1992; 88:249-269. 61. Venkatachalam, CM, Jiang, X, Oldfield, T,.Waldman, M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model 2003; 21:289-307. 62. Pappin, DJ,.Findlay, JBC. Sequence variability in the retinal-attachment domain of mammalian rhodopsins. Biochemical Journal 1984; 217:605613. 63. Bywater, RP, Thomas, D,.Vriend, G. A sequence and structural study of transmembrane helices. J Comput Aided Mol Des 2001; 15:533-552. 64. Dube, P, DeCostanzo, A,.Konopka, JB. Interaction between transmembrane domains five and six of the alpha-factor receptor. J Biol Chem 2000; 275:26492-26499. 65. Ghanouni, P, Steenhuis, JJ, Farrens, DL,.Kobilka, BK. Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc Natl Acad Sci U S A 2001; 98:5997-6002. 66. Javitch, JA, Fu, D, Liapakis, G,.Chen, J. Constitutive activation of the beta2 adrenergic receptor alters the orientation of its sixth membranespanning segment. J Biol Chem 1997; 272:18546-18549. 67. Yeagle, PL,.Albert, AD. A conformational trigger for activation of a G protein by a G protein- coupled receptor. Biochemistry 2003; 42:13651368. 68. Choi, G, Landin, J, Galan, JF, Birge, RR, Albert, AD,.Yeagle, PL. Structural studies of metarhodopsin II, the activated form of the G- protein coupled receptor, rhodopsin. Biochemistry 2002; 41:7318-7324. 69. Angelova, K, Fanelli, F,.Puett, D. A model for constitutive lutropin receptor activation based on molecular simulation and engineered mutations in transmembrane helices 6 and 7. J Biol Chem 2002; 277:32202-32213. 70. Greasley, PJ, Fanelli, F, Scheer, A, Abuin, L, Nenniger-Tosato, M, DeBenedetti, PG,.Cotecchia, S. Mutational and computational analysis of the alpha(1b)-adrenergic receptor. Involvement of basic and hydrophobic residues in receptor activation and G protein coupling. J Biol Chem 2001; 276:46485-46494. 71. Baldwin, JM, Schertler, GF,.Unger, VM. An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol 1997; 272:144-164. 72. Unger, VM, Hargrave, PA, Baldwin, JM,.Schertler, GF. Arrangement of rhodopsin transmembrane alpha-helices. Nature 1997; 389:203-206. 73. Han, M, Lin, SW, Minkova, M, Smith, SO,.Sakmar, TP. Functional interaction of transmembrane helices 3 and 6 in rhodopsin - Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J Biol Chem 1996; 271:32337-32342. 74. Han, M, Lou, JH, Nakanishi, K, Sakmar, TP,.Smith, SO. Partial agonist activity of 11-cis-retinal in rhodopsin mutants. J Biol Chem 1997; 272:23081-23085. 75. Bissantz, C, Bernard, P, Hibert, M,.Rognan, D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins-Structure Function and Genetics 2003; 50:5-25. 76. Jordan, BA, Trapaidze, N, Gomes, I, Nivarthi, R,.Devi, LA. Oligomerization of opioid receptors with beta 2-adrenergic receptors: a role in trafficking and mitogen-activated protein kinase activation. Proc Natl Acad Sci U S A 2001; 98:343-348. 77. McVey, M, Ramsay, D, Kellett, E, Rees, S, Wilson, S, Pope, AJ,.Milligan, G. Monitoring receptor oligomerization using time-resolved fluorescence resonance energy transfer and bioluminescence resonance energy transfer - The human delta-opioid receptor displays constitutive oligomerization at the cell surface, which is not regulated by receptor occupancy. J Biol Chem 2001; 276:14092-14099. 78. Bouvier, M, Moffett, S, Loisel, TP, Mouillac, B, Hebert, T,.Chidiac, P. Palmitoylation of g-protein-coupled receptors - a dynamic modification with functional consequences. Biochem Soc Trans 1995; 23:116-120. 79. Qanbar, R,.Bouvier, M. Role of palmitoylation/depalmitoylation reactions in G-protein- coupled receptor function. Pharmacology & Therapeutics 2003; 97:1-33. 80. ODOWD, BF, HNATOWICH, M, Caron, MG, Lefkowitz, RJ,.Bouvier, M. Palmitoylation of the human beta-2-adrenergic receptor - mutation of Cys341 in the Carboxy tail leads to an uncoupled nonpalmitoylated form of the receptor. J Biol Chem 1989; 264:7564-7569. 81. Adam, L, Bouvier, M,.Jones, TLZ. Nitric oxide modulates beta(2)adrenergic receptor palmitoylation and signaling. J Biol Chem 1999; 274:26337-26343. 82. Bouvier, M, Loisel, TP,.Hebert, T. Dynamic regulation of g-protein coupled receptor palmitoylation - potential role in receptor function. Biochem Soc Trans 1995; 23:577-581. 83. Bouvier, M, Menard, L, Dennis, M,.Marullo, S. Expression and recovery of functional g-protein-coupled receptors using baculovirus expression systems. Current Opinion in Biotechnology 1998; 9:522-527. 84. Yu, HB,.Oprian, DD. Tertiary interactions between transmembrane segments 3 and 5 near the cytoplasmic side of rhodopsin. Biochemistry 1999; 38:12033-12040. 85. Hawtin, SR, Tobin, AB, Patel, S,.Wheatley, M. Palmitoylation of the vasopressin V-1a receptor reveals different conformational requirements for signaling, agonist- induced receptor phosphorylation, and sequestration. J Biol Chem 2001; 276:38139-38146. 86. Hovelmann, S, Hoffmann, SH, Kuhne, R, ter Laak, T, Reilander, H,.Beckers, T. Impact of aromatic residues within transmembrane helix 6 of the human gonadotropin-releasing hormone receptor upon agonist and antagonist binding. Biochemistry 2002; 41:1129-1136. 87. Cotte, N, Balestre, MN, Aumelas, A, Mahe, E, Phalipou, S, Morin, D, Hibert, M, Manning, M, Durroux, T, Barberis, C,.Mouillac, B. Conserved aromatic residues in the transmembrane region VI of the V-1a vasopressin receptor differentiate agonist vs. antagonist ligand binding. Eur J Biochem 2000; 267:4253-4263. 88. Chen, S, Xu, M, Lin, F, Lee, D, Riek, P,.Graham, RM. Phe310 in transmembrane VI of the alpha1B-adrenergic receptor is a key switch residue involved in activation and catecholamine ring aromatic bonding. J Biol Chem 1999; 274:16320-16330. 89. Escrieut, C, Gigoux, V, Archer, E, Verrier, S, Maigret, B, Behrendt, R, Moroder, L, Bignon, E, Silvente-Poirot, S, Pradayrol, L,.Fourmy, D. The biologically crucial C terminus of cholecystokinin and the non-peptide agonist SR-146,131 share a common binding site in the human CCK1 receptor - Evidence for a crucial role of Met- 121 in the activation process. J Biol Chem 2002; 277:7546-7555. 90. Blaker, M, Ren, Y, Seshadri, L, McBride, EW, Beinborn, M,.Kopin, AS. CCK-B/gastrin receptor transmembrane domain mutations selectively alter synthetic agonist efficacy without affecting the activity of endogenous peptides. Mol Pharmacol 2000; 58:399-406. 91. Vogel, WK, Sheehan, DM,.Schimerlik, MI. Site-directed mutagenesis on the m2 muscarinic acetylcholine receptor: The significance of Tyr403 in the binding of agonists and functional coupling. Mol Pharmacol 1997; 52:1087-1094. 92. Mosser, VA, Amana, IJ,.Schimerlik, MI. Kinetic analysis of M-2 muscarinic receptor activation of G(i) in Sf9 insect cell membranes. J Biol Chem 2002; 277:922-931. 93. Wess, J, Maggio, R, PALMER, JR,.Vogel, Z. Role of conserved threonine and tyrosine residues in acetylcholine binding and muscarinic receptor activation - a study with M3-muscarinic-receptor point mutants. J Biol Chem 1992; 267:19313-19319. 94. Karnik, SS, Husain, A,.Graham, RM. Molecular determinants of peptide and non-peptide binding to the AT(1) receptor. Clin Exp Pharmacol Physiol 1996; 23:S58-S66. 95. Nakayama, TA,.Khorana, HG. Mapping of the amino acids in membraneembedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem 1991; 266:4269-4275. 96. Meng, F, Wei, Q, Hoversten, MT, Taylor, LP,.Akil, H. Switching agonist/antagonist properties of opiate alkaloids at the delta opioid receptor using mutations based on the structure of the orphanin FQ receptor. J Biol Chem 2000; 275:21939-21945. 97. Lin, YM, Jian, XY, Lin, ZL, Kroog, GS, Mantey, S, Jensen, RT, Battey, J,.Northup, J. Two amino acids in the sixth transmembrane segment of the mouse gastrin-releasing peptide receptor are important for receptor activation. J Pharmacol Exp Ther 2000; 294:1053-1062. 98. Yang, YK, Dickinson, C, HaskellLuevano, C,.Gantz, I. Molecular basis for the interaction of [Nle(4),D- Phe(7)]melanocyte stimulating hormone with the human melanocortin-1 receptor (melanocyte alpha-MSH receptor). J Biol Chem 1997; 272:23000-23010. 99. Chen, CH, Chen, WY, Liu, HL, Liu, TT, Tsou, AP, Lin, CY, Chao, T, Qi, Y,.Hsiao, KJ. Identification of mutations in the arginine vasopressin receptor 2 gene causing nephrogenic diabetes insipidus in Chinese patients. Journal of Human Genetics 2002; 47:66-73. 100. Berglund, MM, Fredriksson, R, Salaneck, E,.Larhammar, D. Reciprocal mutations of neuropeptide Y receptor Y2 in human and chicken identify amino acids important for antagonist binding. FEBS Lett 2002; 518:5-9. 101. OLAH, ME, REN, HZ, Ostrowski, J, Jacobson, KA,.STILES, GL. Cloning, expression, and characterization of the unique bovine- A1 adenosine receptor - studies on the ligand-binding site by site-directed mutagenesis. J Biol Chem 1992; 267:10764-10770. 102. Mollereau, C, Moisand, C, Butour, JL, Parmentier, M,.Meunier, JC. Replacement of Gln(280) by His in TM6 of the human ORL1 receptor increases affinity but reduces intrinsic activity of opioids. FEBS Lett 1996; 395:17-21. 103. Huang, XP, Nagy, PI, Williams, FE, Peseckis, SM,.Messer, WS. Roles of threonine 192 and asparagine 382 in agonist and antagonist interactions with M-1 muscarinic receptors. Br J Pharmacol 1999; 126:735-745. 104. Wieland, K, Ter Laak, AM, Smit, MJ, Kuhne, R, Timmerman, H,.Leurs, R. Mutational analysis of the antagonist-binding site of the histamine H-1 receptor. J Biol Chem 1999; 274:29994-30000. 105. Fernandez, LM,.Puett, D. Identification of amino acid residues in transmembrane helices VI and VII of the lutropin choriogonadotropin receptor involved in signaling. Biochemistry 1996; 35:3986-3993. 106. Shin, N, Coates, E, Murgolo, NJ, Morse, KL, Bayne, M, Strader, CD,.Monsma, FJ. Molecular Modeling and site-specific mutagenesis of the histamine-binding site of the histamine H-4 receptor. Mol Pharmacol 2002; 62:38-47. 107. Spivak, CE, Beglan, CL, Seidleck, BK, Hirshbein, LD, Blaschak, CJ, Uhl, GR,.Surratt, CK. Naloxone activation of mu-opioid receptors mutated at a histidine residue lining the opioid binding cavity. Mol Pharmacol 1997; 52:983-992. 108. Gao, ZG, Chen, A, Barak, D, Kim, SK, Muller, CE,.Jacobson, KA. Identification by site-directed mutagenesis of residues involved in ligand recognition and activation of the human A(3) adenosine receptor. J Biol Chem 2002; 277:19056-19063. 109. Feighner, SD, Howard, AD, Prendergast, K, Palyha, OC, Hreniuk, DL, Nargund, R, Underwood, D, Tata, JR, Dean, DC, Tan, CP, McKee, KK, Woods, JW, Patchett, AA, Smith, RG,.Van der Ploeg, LHT. Structural requirements for the activation of the human growth hormone secretagogue receptor by peptide and nonpeptide secretagogues. Mol Endocrinol 1998; 12:137-145. 110. Manivet, P, Schneider, B, Smith, JC, Choi, DS, Maroteaux, L, Kellermann, O,.Launay, JM. The serotonin binding site of human and murine 5-HT2B-receptors - Molecular modeling and site-directed mutagenesis. J Biol Chem 2002; 277:17170-17178. 111. Berthold, M, Kahl, U, Jureus, A, Kask, K, Nordvall, G, Langel, U,.Bartfai, T. Mutagenesis and ligand modification studies on galanin binding to its GTP-binding-protein-coupled receptor GalR1. Eur J Biochem 1997; 249:601-606. 112. Jiang, QL, Guo, DP, Lee, BX, VanRhee, AM, Kim, YC, Nicholas, RA, Schachter, JB, Harden, TK,.Jacobson, KA. A mutational analysis of residues essential for ligand recognition at the human P2Y(1) receptor. Mol Pharmacol 1997; 52:499-507. 113. Moro, S, Guo, DP, Camaioni, E, Boyer, JL, Harden, TK,.Jacobson, KA. Human P2Y(1) receptor: Molecular modeling and site-directed mutagenesis as tools to identify agonist and antagonist recognition sites. J Med Chem 1998; 41:1456-1466. 114. Labbe-Jullie, C, Barroso, S, Nicolas-Eteve, D, Reversat, JL, Botto, JM, Mazella, J, Bernassau, JM,.Kitabgi, P. Mutagenesis and modeling of the neurotensin receptor NTR1. Identification of residues that are critical for binding SR 48692, a nonpeptide neurotensin antagonist. J Biol Chem 1998; 273:16351-16357. 115. Greenfeder, S, Cheewatrakoolpong, B, Billah, M, Egan, RW, Keene, E, Murgolo, NJ,.Anthes, JC. The neurokinin-1 and neurokinin-2 receptor binding sites of MDL103,392 differ. Bioorganic & Medicinal Chemistry 1999; 7:2867-2876. 116. Giolitti, A, Cucchi, P, Renzetti, AR, Rotondaro, L, Zappitelli, S,.Maggi, CA. Molecular determinants of peptide and nonpeptide NK-2 receptor antagonists binding sites of the human tachykinin NK-2 receptor by sitedirected mutagenesis. Neuropharmacology 2000; 39:1422-1429. 117. NARDONE, J,.HOGAN, PG. Delineation of a region in the B2 bradykinin receptor that is essential for high-affinity agonist binding. Proc Natl Acad Sci U S A 1994; 91:4417-4421. 118. Ward, SDC, Curtis, CAM,.Hulme, EC. Alanine-scanning mutagenesis of transmembrane domain 6 of the M-1 muscarinic acetylcholine receptor suggests that Tyr381 plays key roles in receptor function. Mol Pharmacol 1999; 56:1031-1041. 119. Javitch, JA, Ballesteros, JA, Weinstein, H,.Chen, J. A cluster of aromatic residues in the sixth membrane-spanning segment of the dopamine D2 receptor is accessible in the binding-site crevice. Biochemistry 1998; 37:998-1006. 120. Akeson, M, Sainz, E, Mantey, SA, Jensen, RT,.Battey, JF. Identification of four amino acids in the gastrin-releasing peptide receptor that are required for high affinity agonist binding. J Biol Chem 1997; 272:1740517409. 121. Gardella, TJ, Luck, MD, Fan, MH,.Lee, CW. Transmembrane residues of the parathyroid hormone (PTH)/PTH- related peptide receptor that specifically affect binding and signaling by agonist ligands. J Biol Chem 1996; 271:12820-12825. 122. Jensen, CJ, Gerard, NP, Schwartz, TW,.Gether, U. The species selectivity of chemically distinct tachykinin nonpeptide antagonists is dependent on common divergent residues of the rat and human neurokinin-1 receptors. Mol Pharmacol 1994; 45:294-299. 123. Sainz, E, Akeson, M, Mantey, SA, Jensen, RT,.Battey, JF. Four amino acid residues are critical for high affinity binding of neuromedin B to the neuromedin B receptor. J Biol Chem 1998; 273:15927-15932. 124. Parker, EM, GRISEL, DA, IBEN, LG,.SHAPIRO, RA. A single amino-acid difference accounts for the pharmacological distinctions between the rat and human 5-hydroxytryptamine 1B receptors. J Neurochem 1993; 60:380-383. 125. Glennon, RA, Dukat, M, Westkaemper, RB, Ismaiel, AM, Izzarelli, DG,.Parker, EM. The binding of propranolol at 5-hydroxytryptamine(1D beta) T355N mutant receptors may involve formation of two hydrogen bonds to asparagine. Mol Pharmacol 1996; 49:198-206. 126. Stitham, J, Stojanovic, A, Merenick, BL, O'Hara, KA,.Hwa, J. The unique ligand-binding pocket for the human prostacyclin receptor - Site-directed mutagenesis and molecular modeling. J Biol Chem 2003; 278:42504257. 127. Dragic, T, Trkola, A, Thompson, DAD, Cormier, EG, Kajumo, FA, Maxwell, E, Lin, SW, Ying, WW, Smith, SO, Sakmar, TP,.Moore, JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci U S A 2000; 97:5639-5644. 128. Waugh, DJJ, Gaivin, RJ, Zuscik, MJ, Gonzalez-Cabrera, P, Ross, SA, Yun, J,.Perez, DM. Phe-308 and Phe-312 in transmembrane domain 7 are major sites of alpha(1)-adrenergic receptor antagonist binding Imidazoline agonists bind like antagonists. J Biol Chem 2001; 276:25366-25371. 129. Suryanarayana, S,.Kobilka, BK. Amino acid substitutions at position 312 in the seventh hydrophobic segment of the beta 2-adrenergic receptor modify ligand-binding specificity. Mol Pharmacol 1993; 44:111-114. 130. Gerber, BO, Meng, EC, Dotsch, V, Baranski, TJ,.Bourne, HR. An activation switch in the ligand binding pocket of the C5a receptor. J Biol Chem 2001; 276:3394-3400. 131. WEITZ, CJ,.Nathans, J. Rhodopsin activation - effects on the metarhodopsin-I metarhodopsin-II equilibrium of neutralization or introduction of charged amino-acids within putative transmembrane segments. Biochemistry 1993; 32:14176-14182. 132. Liang, XY, Parkinson, JA, Weishaupl, M, Gould, RO, Paisey, SJ, Park, HS, Hunter, TM, Blindauer, CA, Parsons, S,.Sadler, PJ. Structure and dynamics of metallomacrocycles: Recognition of zinc xylyl-bicyclam by an HIV coreceptor. Journal of the American Chemical Society 2002; 124:9105-9112. 133. Lundstrom, K, Turpin, MP, Large, C, Robertson, G, Thomas, P,.Lewell, XQ. Mapping of dopamine D-3 receptor binding site by pharmacological characterization of mutants expressed in CHO cells with the Semliki Forest virus system. Journal of Receptor and Signal Transduction Research 1998; 18:133-150. 134. Gouldson, P, Legoux, P, Carillon, C, Delpech, B, Le Fur, G, Ferrara, P,.Shire, D. Contrasting roles of leu(356) in the human CCK(1) receptor for antagonist SR 27897 and agonist SR 146131 binding. Eur J Pharmacol 1999; 383:339-346. 135. Donohue, PJ, Sainz, E, Akeson, M, Kroog, GS, Mantey, SA, Battey, JF, Jensen, RT,.Northup, JK. An aspartate residue at the extracellular boundary of TMII and an arginine residue in TMVII of the gastrinreleasing peptide receptor interact to facilitate heterotrimeric G protein coupling. Biochemistry 1999; 38:9366-9372. 136. Mirzadegan, T, Diehl, F, Ebi, B, Bhakta, S, Polsky, I, McCarley, D, Mulkins, M, Weatherhead, GS, Lapierre, JM, Dankwardt, J, Morgans, D, Wilhelm, R,.Jarnagin, K. Identification of the binding site for a novel class of CCR2b chemokine receptor antagonists - Binding to a common chemokine receptor motif within the helical bundle. J Biol Chem 2000; 275:25562-25571. 137. Lu, ZL, Saldanha, JW,.Hulme, EC. Transmembrane domains 4 and 7 of the M-1 muscarinic acetylcholine receptor are critical for ligand binding and the receptor activation switch. J Biol Chem 2001; 276:34098-34104. 138. Kopin, AS, McBride, EW, Quinn, SM, Kolakowski, LF,.Beinborn, M. The role of the cholecystokinin-B gastrin receptor transmembrane domains in determining affinity for subtype-selective ligands. J Biol Chem 1995; 270:5019-5023. 139. Owens, CE,.Akil, H. Determinants of ligand selectivity at the kappareceptor based on the structure of the orphanin FQ receptor. J Pharmacol Exp Ther 2002; 300:992-999. 140. Meng, F, Taylor, LP, Hoversten, MT, Ueda, Y, Ardati, A, Reinscheid, RK, Monsma, FJ, Watson, SJ, Civelli, O,.Akil, H. Moving from the Orphanin FQ receptor to an opioid receptor using four point mutations. J Biol Chem 1996; 271:32016-32020. 141. Meng, F, Ueda, Y, Hoversten, MT, Taylor, LP, Reinscheid, RK, Monsma, FJ, Watson, SJ, Civelli, O,.Akil, H. Creating a functional opioid alkaloid binding site in the orphanin FQ receptor through site-directed mutagenesis. Mol Pharmacol 1998; 53:772-777. 142. Casarosa, P, Menge, WM, Minisini, R, Otto, C, van Heteren, J, Jongejan, A, Timmerman, H, Moepps, B, Kirchhoff, F, Mertens, T, Smit, MJ,.Leurs, R. Identification of the first nonpeptidergic inverse agonist for a constitutively active viral-encoded G protein-coupled receptor. J Biol Chem 2003; 278:5172-5178. 143. Gouldson, PR, Legoux, P, Carillon, C, Delpech, B, Le Fur, G, Ferrara, P,.Shire, D. Contrasting roles of Leu356 in the human CCK1 receptor for antagonist SR 27897 and agonist SR 146131 binding. Eur J Pharmacol 1999; 383:341-348. 144. Kedzie, KM, Donello, JE, Krauss, HA, Regan, JW,.Gil, DW. A single amino-acid substitution in the EP2 prostaglandin receptor confers responsiveness to prostacyclin analogs. Mol Pharmacol 1998; 54:584590. 145. RIM, J,.Oprian, DD. Constitutive activation of opsin - interaction of mutants with rhodopsin kinase and arrestin. Biochemistry 1995; 34:11938-11945. 146. Dalpiaz, A, Townsend-Nicholson, A, Beukers, MW, Schofield, PR,.Ijzerman, AP. Thermodynamics of full agonist, partial agonist, and antagonist binding to wild-type and mutant adenosine A(1) receptors. Biochem Pharmacol 1998; 56:1437-1445. 147. KIM, JH, Wess, J, VanRhee, AM, Schoneberg, T,.Jacobson, KA. Sitedirected mutagenesis identifies residues involved in ligand recognition in the human A(2A) adenosine receptor. J Biol Chem 1995; 270:1398713997. 148. Townsend-Nicholson, A,.Schofield, PR. A threonine residue in the 7th transmembrane domain of the human A(1)-adenosiner eceptor mediates specific agonist binding. J Biol Chem 1994; 269:2373-2376. 149. Gether, U,.Kobilka, BK. G protein-coupled receptors - ii - mechanism of agonist activation. J Biol Chem 1998; 273:17979-17982. 150. De Lean, A, Stadel, JM,.Lefkowitz, RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclasecoupled beta-adrenergic receptor. J Biol Chem 1980; 255:7108-7117. 151. Krebs, A, Villa, C, Edwards, PC,.Schertler, GFX. Characterisation of an improved two-dimensional p22121 crystal from bovine rhodopsin. J Mol Biol 1998; 282:991-1003. 152. Kjelsberg, MA, Cotecchia, S, Ostrowski, J, Caron, MG,.Lefkowitz, RJ. Constitutive activation of the alpha 1B-adrenergic receptor by all amino acid substitutions at a single site. Evidence for a region which constrains receptor activation. J Biol Chem 1992; 267:1430-1433. 153. Porter, JE, Hwa, J,.Perez, DM. Activation of the alpha1b-adrenergic receptor is initiated by disruption of an interhelical salt bridge constraint. J Biol Chem 1996; 271:28318-28323. 154. Kim, JM, Altenbach, C, Thurmond, RL, Khorana, HG,.Hubbell, WL. Structure and function in rhodopsin: rhodopsin mutants with a neutral amino acid at E134 have a partially activated conformation in the dark state. Proc Natl Acad Sci U S A 1997; 94:14273-14278. 155. Hulme, EC, Lu, ZL, Ward, SD, Allman, K,.Curtis, CA. The conformational switch in 7-transmembrane receptors: the muscarinic receptor paradigm. Eur J Pharmacol 1999; 375:247-260. 156. Perez, DM, Hwa, J, Gaivin, R, Mathur, M, Brown, F,.Graham, RM. Constitutive activation of a single effector pathway: evidence for multiple activation states of a G protein-coupled receptor. Mol Pharmacol 1996; 49:112-122. 157. Teller, DC, Okada, T, Behnke, CA, Palczewski, K,.Stenkamp, RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of g-protein-coupled receptors (gpcrs). Biochemistry 2001; 40:7761-7772. 158. Elling, CE, Nielsen, SM,.Schwartz, TW. Conversion of antagonist-binding site to metal-ion site in the tachykinin nk-1 receptor. Nature 1995; 374:74-77. 159. Beck, M, Sakmar, TP,.Siebert, F. Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry 1998; 37:7630-7639. 160. Gouldson, PR, Winn, PJ,.Reynolds, CA. A molecular dynamics approach to receptor mapping - application to the 5HT3 2-adrenergic receptors. J Med Chem 1995; 38:4080-4086. The effect of ligand promiscuity in the evaluation of GPCR homology models by means of virtual High Throughput Screening Abstract Earlier on (Gouldson et al. 2004) we presented results whereby two distinct β2 adrenergic receptor models were produced using homology modeling based on the structure of bovine rhodopsin and using sets of experimental distance restraints, which characterise active or inactive receptor conformations. The distance restraints were obtained from published data for site-directed cross-linking, engineered zinc binding, site-directed spin-labelling, IR spectroscopy and cysteine accessibility studies conducted on class A GPCRs. The quality of those models was validated with a number of thorough tests as well as a virtual high throughput screening of the two models against a number of GPCR specific ligands. The hypothesis suggested that the ‘active receptor model’ would bind preferentially to β2 specific agonists and the ‘inactive model’ would preferentially bind to β2 specific antagonists over any other GPCR ligands. Here we will show how the enrichment plots obtained as well as the calculated values for the enrichment, yield and hit rates can be affected from ligand promiscuity and the definition of true binders. Introduction vHTS (virtual high-throughput screening) is a very promising computational technique that has proven to save medicinal chemists a substantial amount of time and an even more substantial amount of expenses for pharmaceutical industries. It has been thoroughly used for screening large combinatorial chemistry drug-like molecules or existing drug databases (as described in chapter 4 with the screensaver project) usually against biologically significant protein targets. However, the applications of vHTS are not just limited to screening databases in the hope of finding a lead for a target, vHTS can be used for evaluating how well a binding domain of a protein has been modeled. Bissantz et al (Bissantz et al. 2003) showed that vHTS can be used as means for validating the structure of a GPCR homology model structure. This method of structure validation has several limitations as most of the computational simulations of biological processes are based on approximations and assumptions usually in terms of the energy force fields used as well as the inexistent or very limited protein so that the calculations for these simulations can be possible with the current CPU availability. Other than the problems that are generally associated and well documented with ligand docking (need refs) here we present a new issue with ligand docking as a method of structure validation, ligand promiscuity. In a previous study we presented the enrichment data for two b2 adrenoreceptors. As the availability of crystal structures in the GPCR family is only limited to the dark state (inactive) bovine rhodopsin, this was used as a starting point for the construction of the homology models of the ‘active’ and ‘inactive’ form of the b2 receptor. Methods Need Nicolas’s bit here…What info with regards to docking do I need? Results (I should probably do a t-test here to check for a significance in the difference) 100.00% 90.00% 80.00% True Positive Fraction 70.00% 60.00% Inactive before Active after 50.00% Active before Inactive after 40.00% 30.00% 20.00% 10.00% 0.00% 0.00% 10.00% 20.00% 30.00% 40.00% 50.00% 60.00% False Positive Fraction 70.00% 80.00% 90.00% 100.00% 50 40 30 actives 20 Inactives 10 0 -100 -80 -60 -40 -20 0 20 40 60 80 100 120 140 160 -10 50 40 30 1 actives 20 Inactives 10 0 -100 -80 -60 -40 -20 0 20 40 60 80 100 120 140 -10 Inactive before promiscuity correction Hit list Yield Hit rate Enrichment (%) (%) (%) Factor Obs. Max Obs Max Obs Max 160 Antagonist apoprotein-based model 5 10 15 20 25 4 18 36 50 54 32 57 86 100 100 11 31 42 45 39 100 100 100 100 100 0.4 1.3 1.9 2.1 1.8 4.0 4.1 4.5 4.1 3.3 Inactive after promiscuity correction Hit list Yield Hit rate Enrichment (%) (%) (%) Factor Obs. Max Obs Max Obs Max Antagonist apoprotein-based model 5 10 15 20 25 8 11 22 31 42 19 42 61 78 97 43 27 36 39 42 100 100 100 100 100 1.6 1.1 1.4 1.5 1.7 3.8 4.0 4.0 3.9 3.9 Active before promiscuity correction Hit list Yield Hit rate Enrichment (%) (%) (%) Factor Obs. Max Obs Max Obs Max Agonists (max 16): active model 5 10 15 20 25 38 50 63 88 88 50 100 100 100 100 75 50 42 44 35 100 100 100 100 100 5.7 4.4 4.0 4.1 3.4 7.7 8.7 6.3 4.7 3.9 Active after promiscuity correction Hit list Yield Hit rate Enrichment (%) (%) (%) Factor Obs. Max Obs Max Obs Max Agonists (max 16): active model 5 16 10 35 15 51 20 59 25 73 Discussion 19% 38% 57% 76% 95% 86 93 90 79 79 100 100 100 100 100 3.2 3.4 3.3 2.9 2.9 3.7 3.7 3.7 3.7 3.7 1. Bissantz’s theory that GPCR homology models based on bovine Rhodopsin can be considered as suitable targets for Protein-based virtual Screening is supported by our data 2. The homology models produced are of high quality as shown by the high enrichment curves (especially in the case of the active model) 3. The fact that we used ligands that bind to the same subfamily of proteins (GPCR) as the adrenergic receptor as rogue ligands and not random drug-like ligands meant that some of those ligands may interact with the adrenoreceptors as the GPCRs are meant to have similar binding sites, and this reflected in our study with the… 4. …ligand promiscuity issue which affected the enrichment plots and figures…. Reference List 1. Gouldson,P.R., Kidley,N.J., Bywater,R.P., Psaroudakis,G., Brooks,H.D., Diaz,C., Shire,D., and Reynolds,C.A. 2004. Toward the active conformations of rhodopsin and the beta2-adrenergic receptor. Proteins 56:67-84. 2. Bissantz,C., Bernard,P., Hibert,M., and Rognan,D. 2003. Protein-based virtual screening of chemical databases. II. Are homology models of G-Protein Coupled Receptors suitable targets? Proteins 50:5-25. Comment [G1]:
© Copyright 2026