Distinct Phylogeographic Patterns of Sympatric West Atlantic Exploited Seahorses PADRON, MARIANA Cohen, Sarah
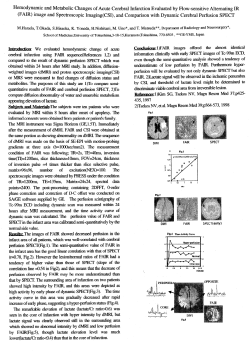
Distinct Phylogeographic Patterns of Sympatric West Atlantic Exploited Seahorses PADRON, 1 Center 1,2 MARIANA Cohen, 2 Sarah and Hamilton, 1 Healy for Applied Biodiversity Informatics, California Academy of Sciences, San Francisco, CA 2 Department of Biology, San Francisco State University, San Francisco, CA . Introduction Results One of the central questions in marine conservation biology is the The results obtained for the mitochondrial DNA indicate that H. extent of population connectivity in marine systems. Several studies erectus and H. reidi exhibit distinct patterns of geographic genetic have addressed this question in the past, with mixed success in structure. An AMOVA revealed significant structure between the resolving genetic structure in populations of marine fishes and populations from the Caribbean and the Atlantic Ocean in both invertebrates from the Indo-Pacific region and Atlantic Ocean. cytochrome b (ФST = 0.737, P ‹ 0.0001; ФCT = 0.690, P ‹ 0.0001) and We are particularly interested in examining the geographic patterns control region (ФST = 0.877, P ‹ 0.0001; ФCT = 0.812, P ‹ 0.0001) for H. of genetic connectivity in two seahorse species with largely reidi; while no significant structure among regions was observed for sympatric distributions: the long-snout seahorse (Hippocampus reidi) H. erectus in both cytochrome b (ФST = 0.220, P ‹ 0.0001; ФCT = 0.006, P and the lined seahorse (Hippocampus erectus). These two exploited › 0.05) and control region (ФST = 0.109, P < 0.05; ФCT = 0.047, P › 0.05). A Fig. 6A & B.- Maximum Likelihood (ML) phylogenetic reconstructions for H. reidi (A) and H. erectus (B) (with outgroups) using GeoPhylobuilder 1.1 Caribbean fish species demonstrate unique life history attributes, that support a test of ecological vs. geographic population structure. N = 38 H = 16 2 B Gene diversity (Hd) High 5 14 3 13 11 Low 16 8 9 4 10 6 15 7 1 12 Honduras A Venezuela B Brazil Photo: Adriana Perez www.flickr.com Fig.1.- Hippocampus erectus Fig.2.- Hippocampus reidi Fig. 4.- Haplotype network of H. reidi (H = 16) 7 Honduras Is there evidence of significant genetic population structure in H. erectus or H. reidi from several populations sampled across their 15 4 29 30 26 Venezuela 43 20 Mexico 23 Can we determine the level of gene flow among populations, or 22 distributed seahorse species across the Caribbean, Gulf of Mexico and 32 Atlantic Ocean. The large break in the distribution of H. reidi from 33 36 37 3 1 38 42 Brazil to Honduras was also supported by reciprocal monophyly in 5 17 our phylogenetic reconstructions. This pattern is consistent with the 41 39 6 31 2 identify areas of high genetic diversity to assess the potential need for distinct management units? We document distinct patterns of genetic variation in two sympatric 27 8 19 high values of molecular divergence between the populations from 18 16 9 12 35 10 34 Brazil and the rest of the Caribbean (Dxy = 0.00857), relative to the 25 24 11 21 molecular divergence between the Caribbean populations of H. reidi 13 14 and its sister species from the Eastern Atlantic, H. algiricus (Dxy = Methods Fig. 5.- Haplotype network of H. erectus (H = 43) A 820 bp fragment of the 0.00677). It seems that the populations of H. reidi from Brazil, Venezuela and Honduras could be treated as distinct management A total of 16 unique haplotypes were found for H. reidi , with no mitochondrial cytochrome b units. Additional sampling of Brazilian populations of H. reidi, as well shared haplotypes between populations. gene and a 285 bp fragment as further comparative analysis with populations of H. algiricus, is For H. erectus, 43 unique haplotypes were identified, with only 2 of the mitochondrial control needed to help resolve taxonomic boundaries in H. reidi. shared haplotypes. One shared between the populations from region were analyzed from On the other hand, H. erectus exhibit no signals of population Florida (82.36%), Honduras (5.88%), Virginia (5.88%) and Colombia Fig.3.- Map of the sampling distribution of H. erectus and H. reidi in the Caribbean and Western Atlantic. individuals of H. reidi from 3 locations of the Caribbean and Western Atlantic. We used phylogenetic reconstructions using PAUP (v. 4.0, Swofford 2002) and MrBayes (v. 3.1, Huelsenbeck and Ronquist 2001), haplotype network with TCS (v. 1.21, Posada and Crandall 2000) and analysis of molecular variance (AMOVA) using Arlequin (v. 3.1, Excoffier et al 2005) to infer population structure within the Atlantic Ocean basin. Five variable microsatellite loci for each species are also being analyzed to estimate population structure and gene flow among populations. 28 Discussion Colombia 40 from 6 locations and 40 N = 72 H = 43 Virginia, US geographic ranges? 70 individuals of H. erectus Fig. 7A & B.- Gene diversity interpolation for H. reidi (A) and H. erectus (B) using ArcMap 9.3 Florida, US Our questions structure between ocean basins, and the haplotype network (5.88%); and another one shared only between the populations from constructed for both the control region and cytochrome b showed a Florida (42.86%) and Mexico (57.14%). high degree of reticulation between the Caribbean and Gulf of Summary of mtDNA for sampled populations of H. erectus and H. reidi from the Western Atlantic Mexico, suggestive of incomplete lineage sorting. H. erectus appears H. erectus Population Venezuela Colombia Mexico Honduras Florida Virginia H. reidi Population Venezuela Honduras Brazil B # of Seq. n 6 13 15 12 24 2 # of Haplotypes H 6 11 10 9 9 2 Gene diversity Hd 1 0.914 0.914 0.909 0.659 1 Nucleotide div π 0.01237 0.00276 0.00300 0.00574 0.00276 0.00385 # of Seq. n 15 13 10 # of Haplotypes H 2 9 5 Gene diversity Hd 0.133 0.936 0.756 Nucleotide div π 0.00024 0.00346 0.00130 to be a panmictic population that could probably be treated as one management unit.. References Mobley, K., et al.2010. Population structure of the dusky pipefish (Syngnathus floridae) from the Atlantic and Gulf of Mexico, as revealed by mitochondrial DNA and microsatellite analyses. Journal of Biogeography 37, 1362-1377. Saarman, N., et al. 2010. Genetic differentiation avvross esatern Pacific oceanographic barriers in the threatened seahorse Hippocampus ingens. Conservation Genetics . Acknowledgement Lakeside Foundation – California Academy of Sciences Scholarship
© Copyright 2026