EXERCISES LRMIX STUDIO
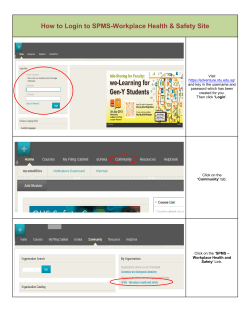
19.04.2015 EXERCISES LRMIX STUDIO EXERCISE 1 – TWO LOCI EXAMPLE In this exercise you will manually create the input files for LRmix Studio. You can use any text editor or spreadsheet software able to save in a compatible file format. I recommend the free and/or open-source software Notepad++ (http://notepad-plus-plus.org/), LibreOffice Calc (http://www.libreoffice.org/), or OpenOffice Calc (http://www.openoffice.org/). Closed source commercial software like Microsoft Excel or Notepad also work. LRmix Studio accepts both comma separated value files (CSV) and tab delimited text files (TXT). Because CSV files can cause problem in countries where semi-colon (;) is used instead of the comma (,) I strongly recommend to use tab delimited text files. Some spread-sheet software by default tries to detect dates and format accordingly (locale dependent). To avoid e.g. 9.3 being changed to 09.mar I recommend using a text editor. If you do not have date problems, or rather use spread-sheet software, remember to save as a tab delimited text file. The correct input files can be found in the TwoLoci folder. a) Create the input file for the DNA profile shown in Figure 1. LRmix Studio can handle the GeneMapperID-X format shown in Table 1 and the LRmix format shown in Table 2. b) Create the input file for the reference profiles as shown in Table 3. c) Create the allele frequency table as shown in Table 4. Note that the sum for each marker equals 1. d) Start a new session of LRmix Studio and import the files you created in task a) - c). e) Analyse the DNA profile using the propositions Hp:V+S and Hd:V+U with dropout probability = 0 for all contributors, drop-in probability = 0, and theta correction = 0. Compare the LRs and overall likelihood ratio with the calculations made by hand. FIGURE 1. A SIMPLE EXAMPLE, WITH ONLY TWO MARKERS, SHOWING A MIXTURE OF TWO PERSONS. Sample Name Marker evidence D3S1358 evidence TH01 Allele 1 Allele 2 Allele 3 Allele 4 15 16 17 18 6 7 8 9.3 TABLE 1. THE GENOTYPES TABLE EXPORTED FROM GENEMAPPERID-X. SampleName Marker evidence D3S1358 evidence TH01 Allele1 Allele2 Allele3 Allele4 15 16 17 18 6 7 8 9.3 TABLE 2. THE LRMIX FORMAT. Page 1 (4) 19.04.2015 Sample Name Marker victim D3S1358 victim TH01 suspect D3S1358 suspect TH01 Allele 1 Allele 2 15 18 8 9.3 16 17 6 7 TABLE 3. THE REFERENCE PROFILES FOR THE VICTIM AND SUSPECT. Allele D3S1358 TH01 5 0.01 6 0.21 7 0.21 8 0.08 9 0.14 9.3 0.34 10 0.01 11 0.01 13 0.01 14 0.12 15 0.27 16 0.23 17 0.20 18 0.14 19 0.02 TABLE 4. APPROXIMATE ALLELE FREQUENCIES FOR D3S1358 AND TH01. EXERCISE 2 – RAPE2 Now we move on to the complete case. The input files can be found in the Rape2 folder. a) Still in LRmix Studio select the Analysis tab and load the allele frequency file esx_norway.txt from the Rape2 folder. Select the Sample Files tab and click the Restart button. NB! If Restart is clicked before loading the new allele frequency file it is not possible to change until after the sample and reference files have been loaded, and the Analysis tab gets activated. If this is the case and the markers sets differ between the previous and new allele frequency files only the markers available in the previous file is checked in the Profile Summary tab. Manually check all markers in the Profile Summary tab before running the analysis. This behaviour applies to LRmix Studio 1.0.2. b) Load the DNA evidence profile stored in the file crime_sample_ldt200.txt. c) Select the Reference Files tab and load the known genotypes stored in the file reference_samples.txt. d) Analyse the DNA profile using the same propositions and settings as in Exercise 1: Hp:V+S and Hd:V+U with dropout probability = 0 for all contributors, drop-in probability = 0, and theta correction = 0. e) What is the overall likelihood ratio? Page 2 (4) 19.04.2015 f) Examine the LR per marker. Which markers are responsible for the overall likelihood ratio? Explain the result. The LR for D3S1358 and TH01 should have similar values as in Exercise 1 (although slightly different allele frequencies are used. g) A model that can handle unexplained peaks is needed here. Examine the EPG. Which approach would you rather take and why? a. Add an extra unknown person in the propositions: Hp:V+S+U and Hd:V+2U b. Allow for drop-in with a probability of e.g. 0.05 c. Remove allele 14 in D10S1248 and allele 23 in FGA because they are most likely artefacts d. Perform an additional PCR of the DNA extract e. Other h) We will continue this exercise with the two-person propositions and a drop-in probability of 0.05. Run the calculation again. How does this change the overall LR and the LR per marker? i) From now on (including other exercises) we will use a theta correction = 0.01. Run the calculation again. How does this change the overall LR and the LR per marker? j) Open the file case_information.txt (double click the file in e.g. Windows Explorer) and read the information about the case. Then take another look at the EPG and consider the risk of drop-out. k) Since this is an intimate sample (i.e. sampled taken from the victim’s body) and all the victim’s alleles are visible and well above the stochastic threshold we can condition upon the victim being present in the mixture. We do this by setting the drop-out probability for the victim to 0 under both hypotheses in the Analysis tab. l) What about the drop-out probability for the suspect and unknown? All alleles from the suspect are visible (although some are below the stochastic threshold) so we expect the drop-out probability to be low. For the unknown contributors we really don’t know since the genotype is unknown. Fortunately LRmix Studio can help estimate the drop-out probability range by simulation. Click the Sensitivity Analysis tab. We want to vary the drop-out probability for the suspect and unknown so accept the default and click Run in both the Sensitivity Analysis Settings tab and the Dropout Estimation Settings tab (it does not matter which one you run first). A range of plausible drop-out values are marked in the plot. Always check the EPG to see that the range is reasonable. Look at the LR curve and note the most conservative drop-out value. m) Go back to the Analysis tab and plug in the conservative drop-out value for the suspect and unknown. Click Run to calculate the new LR. How does this change the overall LR and the LR per marker? n) Click the Non-contributor Test tab. Check the suspect as Person of Interest (the person to be replaced by random profiles). Click Run. o) How would you report this case? Create a report from the analyses and type your conclusion in the remark textbox dialog. p) Examine the report. What is special about allele 9 in D10S1248 for this case? Page 3 (4) 19.04.2015 EXERCISE 3* – MAJOR / MINOR Until now we have observed all reference alleles in the evidence profile. In this exercise the DNA evidence is low-template and multiple drop-outs have occurred. The input files and solution can be found in the MajorMinor folder. EXERCISE 4* – TWO SUSPECTS So far we have only considered one suspect at a time in the propositions. In this exercise you will explore a case with two suspects. The input files and solution can be found in the TwoSuspects (Case 1) folder. EXERCISE 5 – MURDER In this exercise you will explore three-person mixtures from a brutal murder case with two female suspects. One suspect’s reference profile is old and analysed with fewer markers. The input files can be found in the Murder folder. Load the sample and reference files (crime_samples.txt and reference_samples.txt). Read the background information provided in the file case_information.txt. The EPGs are provided in epg1.png, epg2.png, and epg3.png. EXERCISE 6 – SEXUAL ABUSE In this exercise you encounter a case with at least three contributors to the evidence mixture. The input files can be found in the SexualAbuse folder. Load the sample and reference files (crime_samples.txt and reference_samples.txt). Read the background information provided in the file case_information.txt. The EPG is provided in epg.png. Open the Profile Summary tab and examine the evidence and the reference profiles. What is your conclusion? Would you continue the analysis in LRmix Studio? EXERCISE 7* – MULTIPLE SUSPECTS In this exercise you encounter a complex murder case with a male victim killed in a fight. There are five suspects that are apprehended by police and DNA profiled. Is there evidence that any of the suspects’ DNA is at the crime-scene? The input files and solution can be found in the MultipleSuspects folder. EXERCISE 8* – COMPLEX THREE PERSON CASE In this exercise you encounter a complex three person case where a victim is stabbed in the suspect’s flat. The input files and solution can be found in the ComplextThreePersonCase (Case 3) folder. Page 4 (4)
© Copyright 2026