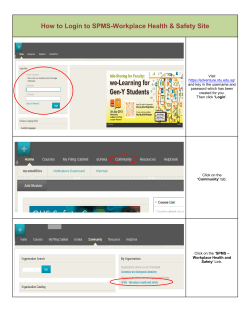
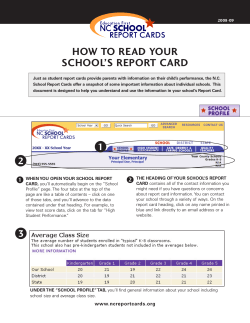
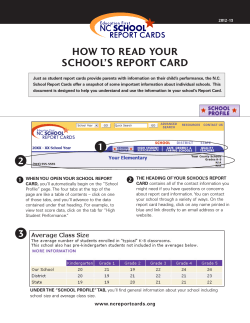
Introduction to R with hands-on exercises
Introduction to R
Mikkel Meyer Andersen
Department of Mathematical Sciences
Aalborg University
Denmark
Apr 21, 2015
About me
I
I
Assistant professor in applied statistics
Main research focus: Evidential weight of lineage markers (so far:
Y-STR) – more will come after this introduction to R
Prerequisites
I
I
R
(RStudio?)
What is R?
Advanced (and basic!) calculator
2 + 2
## [1] 4
cos(2*pi/3)
## [1] -0.5
Graph illustrator
10
0
−10
−20
−30
x^3 + 3 * x^2 − 6 * x − 8
20
30
curve(x^3 + 3*x^2 - 6*x - 8, from = -5, to = 3)
−4
−2
0
x
2
Vectors
1:10
##
[1]
1
2
3
4
c(1, 5, 6)
## [1] 1 5 6
c(1:3, 6:8)
## [1] 1 2 3 6 7 8
5
6
7
8
9 10
Matrices
A <- matrix(1:9, nrow = 3)
A
##
[,1] [,2] [,3]
## [1,]
1
4
7
## [2,]
2
5
8
## [3,]
3
6
9
A[2,2] <- 1000
A
##
[,1] [,2] [,3]
## [1,]
1
4
7
## [2,]
2 1000
8
## [3,]
3
6
9
Programming language
x <- 0
for (i in 1:10) {
x <- x + i
}
x
## [1] 55
sum(1:10)
## [1] 55
double_number <- function(x) 2*x
double_number(4)
## [1] 8
Statistical tool
head(cars)
##
##
##
##
##
##
##
1
2
3
4
5
6
speed dist
4
2
4
10
7
4
7
22
8
16
9
10
Statistical tool
0
20
40
dist
60
80
100
120
plot(dist ~ speed, cars)
5
10
15
speed
20
25
Statistical tool
fit <- lm(dist ~ speed, cars)
summary(fit)
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
Call:
lm(formula = dist ~ speed, data = cars)
Residuals:
Min
1Q
-29.069 -9.525
Median
-2.272
3Q
9.215
Max
43.201
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -17.5791
6.7584 -2.601
0.0123 *
speed
3.9324
0.4155
9.464 1.49e-12 ***
--Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' '
Residual standard error: 15.38 on 48 degrees of freedom
Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
Statistical tool
60
0
20
40
dist
80
100
120
plot(dist ~ speed, cars)
lines(cars$speed, fit$fitted.values)
5
10
15
speed
#abline(a = coef(fit)[1], b = coef(fit)[2])
20
25
Combining features
a <- sample(1:10)
a
##
[1]
3 10
8
5
9
1
6
2
4
7
1
2
9
4
8
7
5
2 10
7
9
3
6
5
1
a[1]
## [1] 3
a[-(1:5)]
## [1] 1 6 2 4 7
sample(1:10)
##
[1]
6
3 10
sample(1:10)
##
[1]
8
4
Extensible
A lot of packages (or libraries) exist
Reproducible documents
Reproducible documents (reports, presentations, etc.): Rmarkdown.
Demonstration later.
Is R difficult?
Yes. The learning curve can be steep. So is talking, we still do the climb.
My usage
I
I
Research (simulation studies)
Reproducible research and reports – yes, “final” datasets do change
Hands-on
Install packages
R is extensible – install a package called disclapmix
install.packages("disclapmix") or RStudio: Tools -> Install
Packages
Search for help
I
I
I
I
Know function name: ?sum
Does not know function name: help.search("search phrase")
Cheat sheet (e.g. http:
//cran.r-project.org/doc/contrib/Short-refcard.pdf)
http://tryr.codeschool.com
Good practises
I
Save your commands in script files (also see History tab in RStudio)
Loading data
I
I
I
I
Loading Excel files can be difficult to set-up (but can be done)
Use Excel to collect, edit, prepare etc. data, then export to CSV (I
prefer tab as field delimiter, others comma)
Import CSV in R
Data cleaning can be time consuming
Example
Supplementary Table 2 from paper: http://www.sciencedirect.com/
science/article/pii/S0379073805000095 (note the table caption is
included – remove)
RStudio: Tools -> Import Dataset -> From text file (remember
checking input, options, and output – e.g. heading)
(Command can be saved to script file for future automated loading.)
View dataset
RStudio:
View(den)
Simple R:
head(den)
##
##
##
##
##
##
##
##
##
##
##
##
##
1
2
3
4
5
6
1
2
3
4
5
Ht DYS19 DYS385a.b DYS389.I DYS389.II DYS390 DYS391 DYS392
1
13
13–16
13
29
22
10
15
2
13
13–19
13
30
25
10
14
3
13
14–23
11
27
23
11
14
4
13
16
14
32
24
10
11
5
13
16–18
13
30
24
10
11
6
14
10–15
13
30
23
11
13
DYS438 DYS439 N Freq.
11
12 1 0.0054
12
12 1 0.0054
12
12 1 0.0054
10
12 1 0.0054
10
12 2 0.0108
Descriptive statistics
sum(den$N)
## [1] 185
dys19 <- rep(den$DYS19, den$N)
table(dys19)
## dys19
## 13 14
##
6 113
15
45
16
18
17
3
summary(dys19)
##
##
Min. 1st Qu.
13.00
14.00
Median
14.00
Mean 3rd Qu.
14.45
15.00
Max.
17.00
Locus dependency
tab <- xtabs(N ~ DYS19 + DYS390, den)
tab
##
DYS390
## DYS19 21 22 23 24 25 26
##
13 0 1 1 3 1 0
##
14 1 30 41 35 4 2
##
15 0 8 9 12 12 4
##
16 0 1 3 4 9 1
##
17 0 0 0 0 3 0
chisq.test(tab)
## Warning in chisq.test(tab): Chi-squared approximation may be
##
## Pearson's Chi-squared test
##
## data: tab
## X-squared = 58.8064, df = 20, p-value = 1.088e-05
Plotting
Histogram:
hist(dys19)
60
40
20
0
Frequency
80
100
Histogram of dys19
13
14
15
16
17
Plotting
Bar plot:
60
40
20
0
table(dys19)
80
100
plot(table(dys19))
13
14
15
dys19
16
17
Rmarkdown
RStudio
File -> New File -> R Markdown
Tips and tricks
Tips and tricks
I
I
I
I
I
I
Horn of plenty: overwhelming functionality (logistic regression,
support vector machines, random forests, . . . )
Print cheat sheet (e.g. http:
//cran.r-project.org/doc/contrib/Short-refcard.pdf)
Plotting: ggplot2
Data cleaning and wrangling: tidyr and dplyr
Questions? R help, Google (R sort data frame) or Stack overflow
at http://stackoverflow.com/questions/tagged/r
RStudio Support about learning R: https://support.rstudio.
com/hc/en-us/categories/200098757-Learn-R
© Copyright 2026