Document 267466
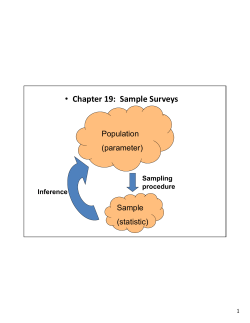
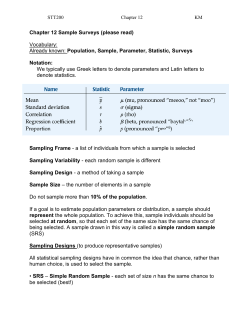
Sampling and sample storage Gemma Henderson (AgResearch, New Zealand) With contributions from: Sandra Kittelmann, Faith Cox, Garry Waghorn, Stefan Muetzel, Xuezhao Sun, Michelle Kirk, Michael Tavendale, Vahideh Heidarian Miri, Michael Zethof, Sarah Lewis, Sam Noel & Peter Janssen & Ilma Tapio (MTT, Finland) With contributions from: Katerina Fliegerova, Aurelie Bonin, Marialuisa Callegari, Jakub Mrazek, Frederic Boyer, Pierre Taberlet, Sara Federici, Paolo Bani, Kevin Shingfield & John Wallace Joint RuminOmics and RMG Network Workshop 22/06/2013, Dublin, Ireland Sampling and storage options • Morning feeding (-1h to +6h) • Evening feeding • Morning/evening combined Timing Feces Sampling type Regurgitated sample Sample fraction Buccal swabs DNA extraction Storage Non invasive methods =(?) Rumen sampling • Stomach tubing • Rumen fistula • At slaughter • Sampling site within the rumen • Liquids • Solids • Mixed rumen content • Frozen (-20C, -80C, N2) • Freeze-dried • Mixed with cryo-protectants (ethanol, RNA later, formalin, PBS/glycerol) Percentage of the community Timing Before v after feeding 0.002 0.009 * * 0.043 * * 0.038 <0.001 Henderson, Sun et al., unpublished * * pasture-fed sheep, n = 10, paired t-test Fistula v stomach tube Percentage of the community Sampling type Henderson et al., PLOS ONE (accepted) * * * ryegrass-fed cows, n = 14, paired t-test Whole v liquid v solid Percentage of the community Sample fraction Henderson et al., PLOS ONE (accepted) * * * * * ryegrass-fed cows, n = 14, paired t-test 0.001 0.035 0.041 Percentage of the community Percentage of the community Percentage of the community Freeze-dried v frozen only Storage 0.002 0.011 ** * * * 0.017 0.008 0.009 0.002 * 0.003 0.005 * * 0.007 Muetzel, Henderson et al., unpublished <0.001 0.041 * * lucerne chaff-fed sheep, n = 6, paired t-test Storage Cryo-protectants In ethanol at RT Without cryo-protectant in -80C In PBS-glycerol in -80C Regurgutated in -80C Without cryo-protectant in -80C Regurgitated in -80C Without cryo-protectant fresh Regurgitated fresh Without cryo-protectant fresh Regurgitated fresh Regurgitated PBS-glycerol ETOH Just frozen -80C -80C RT -80C McKain et al. short communication Animal 1 Animal 2 sheep, n=2 Feces Non-invasive sample collection Families within the Firmicutes Families within the Bacteroidetes de Oliveira et al. 2013 Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Veterinary Microbiology 164: 307-314. DNA extraction Percentage of the community DNA extraction Henderson et al., PLOS ONE (accepted) Take home message Every sampling technique will represent the rumen microbial community It is not yet possible to tell which techniques truly represent the rumen microbial community • There is no “right” or “wrong” Design your study carefully and be aware of differences introduced by selection of even subtly different protocols • ...especially if looking for subtle changes Large cohort studies should strive to use the same or comparable protocols • Global Rumen Census • RuminOmics • ring test Many more downstream variables to be considered • Primers, data analysis, taxonomic frameworks, etc. Acknowledgements New Zealand Sandra Kittelmann Faith Cox Garry Waghorn Stefan Muetzel Xuezhao Sun Michelle Kirk Michael Tavendale Vahideh Heidarian Miri Michael Zethof Sarah Lewis Sam Noel Peter Janssen RuminOmics consortium Katerina Fliegerova Jakub Mrazek Marialuisa Callegari Paolo Bani Sara Federici Aurelie Bonin Frederic Boyer Pierre Taberlet Kevin Shingfield John Wallace Funding PGgRc Ministry for Primary Industries SLMACC Dairy NZ AgResearch Funding RuminOmics Thanks! Other valuable references: Callaway et al. 2010, J Anim Sci 88:3977 De Oliveira et al. 2013, Vet Microbiology 164:307 Dowd et al. 2008, BMC Microbiology 8:125 Fouts et al. 2012, PLOS ONE 7: e48289 Leedle et al. 1982, AEM 44:402 Li et al. 2009, J Appl Microbiol 107:1924 Li et al. 2012, Vet Microbiology 155:72 Lodge-Ivey et al. 2009, J Anim Sci 87: 2333 Prates et al. 2010, Anim Feed Sci Technol 155:186 Rosewarne et al. 2011, Microb Ecol 61:448 Shanks et al. 2011, Appl Environ Microbiol 77:2992 Wu et al. 2010, BMC Microbiology 10:206 …
© Copyright 2026