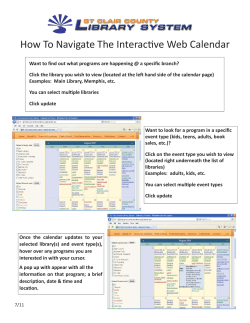
Cellecta Pooled Barcoded Lentiviral shRNA Libraries User Manual
Cellecta Pooled Barcoded Lentiviral shRNA Libraries HT RNAi Genetic Screens with DECIPHER™ or Custom Pooled shRNA Libraries User Manual v5f, 7-3-2014 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Table of Contents A. Background 3 B. Required Materials 4 C. Packaging Protocol for Pooled Lentiviral shRNA Libraries 7 D. Transduction Protocols and Lentiviral Titer Estimation 9 E. Protocols for Genetic Screens with Pooled shRNA Libraries 12 F. Genomic DNA Extraction for Barcode Amplification and HT Sequencing 15 G. Amplification of shRNA-specific Barcodes from Genomic DNA 16 H. HT sequencing of Pooled shRNA-specific Barcodes on Illumina’s HT Sequencing Platform 18 I. Barcode Enumeration (Conversion of raw HT Seq data to number of reads for each barcode) 18 J. Statistical Analysis of shRNA hits enriched/depleted in genetic screen 19 K. Troubleshooting 19 L. Technical Support 21 M. Safety Guidelines 22 N. References 23 O. Appendix 30 P. 1. Lentiviral shRNA Expression Vector Maps 30 2. HT Sequencing Primers 31 3. Common Library Vector Features 32 4. DECIPHER Library HT Sequencing Q.C. Data 33 5. DECIPHER Library Individual Clone Sequencing Q.C. Data 33 6. DECIPHER Project Resources 33 Terms and Conditions 34 Important User Manual Updates from Version 5b A. Background • Human Modules 2 and 3 cannot be combined due to significant barcode sequence overlap. B. Required Materials • We don’t support sequencing on the MiSeq. C. Packaging Protocol for DECIPHER Pooled Lentiviral shRNA Libraries • Day 0 – Plate Cells E. Protocols for Genetic Screens with Pooled shRNA Libraries • For most RNAi screens, we recommend optimizing transduction conditions and performing genetic screen transductions at no more than 0.5 MOI (40% transduction efficiency). • Example of a Negative Selection Viability “Drop out” RNAi Screen (Day 3) H. HT sequencing of Pooled shRNA-specific Barcodes on Illumina’s HT Sequencing Platform • Added modified Illumina HT Sequencing protocol. [email protected] 2 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net A. Background The protocols below provide the instructions on how to package, titer, and transduce target cells with DECIPHER™ or custom pooled lentiviral shRNA libraries. Also provided are examples for both positive and negative selection screens. Additional protocols provide guidelines for the preparation of barcoded probes for high-throughput (HT) sequencing and analysis of raw sequencing data sets. Please read the entire user manual before proceeding with your experiment. The protocols and methods apply specifically to the following DECIPHER Modules, although the protocols are scalable and can be used for smaller or larger libraries. To ensure you have the latest version of this user manual, please visit http://www.cellecta.com/resources/protocols/. DECIPHER Pooled shRNA Library Modules Library Human Module 1 Human Module 2 Human Module 3 Mouse Module 1 Mouse Module 2 Vector pRSI12 pRSI12 pRSI12 pRSI12 pRSI12 Target Genes Signaling Pathways Disease-Associated Cell Surface, Extracellular, DNA Binding Signaling Pathways Disease-Associated # mRNA 5,043 5,412 4,922 4,625 4,520 # shRNA 27,500 27,500 27,500 27,500 27,500 Catalog # DHPAC-M1-P DHDAC-M2-P DHCSC-M3-P DMPAC-M1V2-P DMDAC-M2V2-P IMPORTANT: The barcode sequences in Human Modules 2 and 3 have significant overlap, therefore these modules cannot be combined in any step of the procedure including HT Sequencing. NOTE: The module names are used solely for convenience to describe the major groups of genes targeted in the module. Many genes targeted in a module do not fall within the description, all modules target a variety of genes throughout the genome, and not all genes generally considered to fall under a specific description will be found in the module with the specific gene description. Please refer to the gene lists and complete gene annotations (available on the DECIPHER website at http://www.decipherproject.net/support/#gene-lists) associated with each module for detailed information regarding which genes are present in each specific module. Also, each module targets an orthogonal set of mRNA transcripts so there is no overlap in the targets between modules. Human Genome-Wide Pooled shRNA Library Modules Library hGW Module 1 hGW Module 2 hGW Module 3 hGW Modules 1,2,3 (mixed) Vector pRSI16-13kCB18 pRSI16-13kCB18 pRSI16-13kCB18 pRSI16-13kCB18 # mRNA 6,316 6,316 6,635 19,267 # shRNA 50,469 50,405 52,926 153,800 Plasmid Cat.# HGW-M1-P2 HGW-M2-P2 HGW-M3-P2 HGW-P2 Virus Cat.# HGW-M1-V HGW-M2-V HGW-M3-V HGW-V Designing and Performing HT RNAi Genetic Screens Specific screening protocols will vary depending on the particular biological mechanism to be studied. For general information and examples of successful genetic screening experiments, we recommend that you refer to the References Section. Although the specific protocol and controls may be different depending on the cell type, functional assay, and selection protocol (e.g., FACS, apoptosis induction, toxic chemical survival, etc.), it is critical to carefully design your experiment in order to generate statistically significant data. With this in mind, consider the following suggestions when setting up your experiment: Model Phenotype Selection with Positive Control shRNA Construct(s). Before performing a large-scale genetic screen with a pooled lentiviral shRNA library, we suggest making several shRNA constructs designed against one or more particular target genes whose inactivation is known to elicit the desirable phenotypic changes in the target cells. Then, by packaging and transducing these positive control shRNA constructs into target cells, you can optimize the protocol for enrichment/depletion of cells with induced phenotypic changes for your experiment with the shRNA library. As negative control cells, which should not be enriched/depleted during the [email protected] 3 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net selection steps, you can use cells transduced by non-targeting shRNA constructs (e.g. against Luciferase, or a scrambled shRNA control). Optimize the enrichment/depletion protocol. The quality of genetic screen data will depend significantly on the design and conditions used for the phenotype-specific selection step. A high enrichment/depletion level of target cells with a specific trait will help to identify shRNA constructs that are significantly enriched/depleted above the background noise level of non-enriched/depleted shRNA inserts. In most cases, transduced cells can be used to start a phenotypic screen at approximately 3 days after transduction. However, this is based on anecdotal observations and the time it usually takes the lentiviral cassette to integrate and the shRNAs to express in most cell types. For certain genes and selections, the knockdown effect may happen quickly (2 days) or take significantly longer (4-7 days) to manifest. When performing a genetic screen experiment, make an effort to minimize the time necessary for functional selection. Extended growth of phenotypically-selected cells reduces the reproducibility of identification of functional shRNAs in triplicate cell samples due to heterogeneity of cellular pools, differences in clonal cell growth, spontaneous apoptosis, etc. (i.e. “genetic drift”). Based on our experience, the maximum number of cell duplications for negative selection (viability) screens should be between 8-12 divisions. In the case of positive selection screens with high levels of enrichment (50-100-fold) of phenotypespecific cells, selection should be applied as early as possible after shRNA transduction, and cells should be harvested soon after selection. All harvested cells should be used for purification of genomic DNA and barcode amplification. For positive selection screens with low levels of enrichment (3-10fold), consider designing an experiment with two sequential rounds of enrichment and using the entire pool of second-round enriched cells for genomic DNA isolation and barcode amplification steps. Use Reference Control Cells. As a control for the genetic screen, it may be necessary to use cells infected with the shRNA library but not selected for a specific phenotype or induced (treated) by a phenotype-inducing agent, etc. There are many options that can be considered for the selection of appropriate reference control, depending on your biological system. This control is necessary to use as a standard to measure the relative levels of each shRNA insert species in the transduced cell population without selection. Without an appropriate transduced cell control, it might be difficult or impossible to determine which shRNA species are enriched/depleted in the transduced cells after the selection step, based only on shRNA representation in plasmid library. Design the experiment with at least triplicate samples. In order to achieve statistically significant identification of genes involved in phenotypic responses, in most cases it is necessary to design the experiment with at least triplicate samples for each population of phenotype-selected and reference control cells. B. Required Materials: DECIPHER Project Users: The following components are included with each DECIPHER shRNA Library order: • 120 µg of each plasmid library ordered, in the pRSI12-U6-(sh)-HTS4-UbiC-TagRFP-2A-Puro vector; enough to generate lentivirus for approximately 50-100 screens (depending on cell type) • 10 µg empty library vector, as a packaging and transduction control; or, after linearization by BbsI/BpiI restriction digest, for cloning individual constructs used to validate hits from your screen • User Manual and Product Analysis Certificates (http://www.cellecta.com/resources/protocols/) • List of shRNA and barcode sequences (http://www.decipherproject.net/support/#sequences) • HT Sequencing QC data of plasmid library (http://www.decipherproject.net/support/#sequences) Libraries can be packaged into lentiviral particles with nearly any 2nd or 3rd generation HIV-based lentiviral packaging mix. The vector map, sequence, feature map, and restriction map can be downloaded from the DECIPHER Project website at http://www.decipherproject.net/support/#vector-info. [email protected] 4 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net The following DECIPHER custom services are available from Cellecta at additional cost. For more information, visit www.cellecta.com, email us at [email protected], or call +1-650-938-3910. Additional Products and Services Ready-to-Use Packaging Plasmid Mix (250 µg) DECIPHER Module Packaging (2 × 10^8 TU or 1 × 10^9 TU per module) HT Sequencing of DECIPHER Library experimental samples (frozen cells, DNA, or xenograft) Pre-made or Custom Lentiviral shRNA Constructs (Plasmid or Packaged) Cloning DECIPHER Module into Custom shRNA library vector Catalog # CPCP-K2A CLVP-2E8, CLVP-LGLIB CANA-SQ, CANA-SQD, CANA-SQT many DCLN-M-P Note on The DECIPHER Project: Plasmid DECIPHER shRNA Libraries and vector are distributed free of charge to academic and nonprofit users under a Material Transfer Agreement (MTA). The MTA and Order Form can be downloaded from http://www.decipherproject.net/support/. All Custom Pooled shRNA Library Users: The following deliverables are included or available with each Custom shRNA Library order: • Plasmid and/or packaged library (titers, volumes, and number of tubes may vary between lots) • Empty library vector (with Plasmid library orders only) • Product Analysis Certificate, which includes descriptions of components and individual clone sequencing QC (http://www.cellecta.com/resources/protocols/) • HT Seq QC data of plasmid library (DECIPHER: http://decipherproject.net/support/#sequences) • Sequences of shRNA, barcodes (DECIPHER: http://www.decipherproject.net/support/#sequences) • Library cloning vector map and sequence (http://www.cellecta.com/resources/vectors/) • User Manual (http://www.cellecta.com/resources/protocols/) Packaging of Library Pooled shRNA Libraries can be packaged into lentiviral particles with nearly any 2nd or 3rd generation HIV-based lentiviral packaging mix, or with Cellecta’s Ready-to-Use Packaging Plasmid Mix (Cat.# CPCP-K2A). Cellecta can also provide libraries in ready-to-use packaged format. Please contact us at [email protected] for a custom quote. Materials Available Separately from Cellecta or other vendors: • Positive control (targeting) lentiviral shRNA constructs (Custom or premade, from Cellecta; or generated by customer) • Negative control (non-targeting) lentiviral shRNA constructs (Custom or premade, from Cellecta; or generated by customer) • Linearized shRNA expression vector, for cloning individual constructs used to validate hits from your screen (available from Cellecta) • Lentiviral Packaging Plasmid mix (recommended: mix of packaging plasmids, psPAX2 and pMD2.G, available from AddGene, Cat.#s 12260 and 12259; or custom ordered through Cellecta, Ready-to-Use Lentiviral Packaging Plasmid Mix, in a w:w ratio of 4:1 psPAX2:pMD2.G, Cat.# CPCP-K2A) NOTE: Cellecta’s Packaging Mix is purified by double CsCl gradient. In our experience, CsClpurified plasmid DNA provides higher and more consistent packaging efficiencies than plasmid DNA purified by anion-exchange column or other method. • 293T/17 Cell Line (ATCC, Cat.# CRL-11268™) • Dulbecco's Modified Eagle Medium (D-MEM) (1X) (Mediatech CellGro, Cat.# 15-013-CV) [email protected] 5 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net NOTE: ADD FRESH GLUTAMINE (1X) at the time a sealed bottle of D-MEM is opened, even if the label indicates glutamine has already been added. Glutamine in solution at +4°C has a half-life of 1–2 months, so glutamine(+) D-MEM purchased “off-the-shelf” from a supplier is to be regarded as glutamine(-). In our experience, the addition of glutamine increases titer approximately 2fold. If D-MEM comes supplemented with stable L-Alanyl-L-Glutamine dipeptide, addition of fresh glutamine is not necessary. • Glutamine (L-Alanyl-L-Glutamine, Dipeptide L-glutamine) (Mediatech, Cat.# 25-015-CI) • Fetal Bovine Serum (recommended: Mediatech, Cat.# MT 35-010-CV) • Puromycin • D-PBS (Mediatech, Cat. # 21-031-CV) • Trypsin-EDTA (Mediatech, Cat. # 15-040-CV) • Polybrene® (hexadimethrine bromide) (Sigma-Aldrich, Cat.# 107689) • 500 ml, 0.2 µm filter units (Fisher Scientific Cat.# 09-741-05 or Thermo Scientific Cat.# 5690020) • Tissue Culture Plates and Related Tissue Culture Supplies • Lipofectamine™ Reagent (Life Technologies, Cat.# 18324-020) • Plus™ Reagent (Life Technologies, Cat.# 11514-015) • 15-ml BD FALCON screw-cap centrifuge tubes (12,000 RCF rated, PP, P:CHCl3-resistant, BD Biosciences, Cat.# 352196) • Buffer P1 (50mM Tris-HCl pH 8.0, 10mM EDTA) (QIAGEN, Cat.# 19051) • RNase A (QIAGEN, Cat.# 19101) • Sonicator for Genomic DNA Shearing • Phenol:Chloroform pH 8.0 (Sigma-Aldrich, Cat.# P3803) • DNase I, RNase-free (Epicentre, Cat. D9905K) • Titanium Taq DNA polymerase with PCR buffer (Clontech-Takara, Cat.# 639242) • dNTP Mix (10 mM each) (GE Healthcare, Cat. # 28-4065-52) • QIAquick PCR purification kit (QIAGEN, Cat.# 28106) • QIAquick Gel Extraction Kit (QIAGEN, Cat.# 28706) • Primer for sequencing shRNA inserts in shRNA constructs (IDT)*: See Appendix P.2 • PCR primers for barcode amplification from genomic DNA (IDT)*: See Appendix P.2 • HT sequencing primers (IDT)*: See Appendix P.2 • HT Sequencing Kits (Illumina): Platform Kit Type Illumina Cat.# Description Sequencing FC-104-5001 TruSeq SBS Kit v5 – GA (36-cycle) GAIIx Cluster Generation GD-203-5001 TruSeq SR Cluster Kit v5 – CS – GA Sequencing FC-401-3002 TruSeq SBS Kit v3 – HS (50 cycle) HiSeq* Cluster Generation GD-401-3001 TruSeq SR Cluster Kit v3-cbot-HS * See Illumina website for information on HiSeq 2500 rapid run kits. NOTE: We currently do not support HT sequencing of samples on the Illumina MiSeq. Related Services from Cellecta • Custom Pooled shRNA Library Construction • RNAi Functional Genetic Screens with Pooled shRNA Libraries, Cat.# CRGS-X • HT Barcode Sequencing of Cell Pellets, DNA, or Xenografts from RNAi Screen (with Cellecta Library) • Pre-made and Custom shRNA and CRISPR Constructs • Linearized shRNA Expression Vectors [email protected] 6 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net C. Packaging Protocol for Pooled Lentiviral shRNA Libraries The following protocol describes the generation of a packaged DECIPHER pooled lentiviral 27K shRNA library (27K shRNA complexity) using Invitrogen’s Lipofectamine™ and Plus™ Reagent (see Required Materials). Other transfection reagents may be used, but the protocol should be adjusted to fit the manufacturer’s protocol. The yield of recombinant lentiviral particles typically produced under these optimized conditions is 1-10 × 106 TU/ml. In this protocol, using ten (10) 15-cm plates, at least 3 × 108 TU of total lentiviral particles can be produced. 1. Start growing 293T cells in D-MEM medium plus glutamine (see Required Materials), supplemented with 10% FBS without antibiotics, 2 to 3 days prior to transfection. Day 0 – Plate Cells 2. Twenty four (24) hours prior to transfection, plate 12.5 × 106 293T cells in each of ten (10) 15-cm plates (or 150 cm2 flasks). Use 30 ml of media per plate. Disperse the cells and ensure even distribution. At the moment of transfection, the cells should have reached ~80% confluency. Increase or decrease the number of 293T cells seeded if optimal confluency is not achieved in 24 hours. Incubate at 37°C in a CO2 incubator for 24 hours. Day 1 – Transfection (Ten 15-cm plates*) 3. In sterile 50-ml polypropylene tube, mix 600 µl (300 µg) of the Ready-to-use Packaging plasmid mix (see Required Materials for formulation) with 60 µg** of the plasmid library and add the plasmid mixture to 12 ml D-MEM medium without serum or antibiotics. Add 600 µl of Plus Reagent, mix, and incubate at room temperature for 15 min. 10X 15-cm plates 600 µl 60 µl 12,000 µl 600 µl 13,260 µl Component Ready-to-use Packaging Plasmid Mix (0.5 µg/µl) * Plasmid shRNA Library (1 µg/µl) ** D-MEM, no FBS, no antibiotics Plus Reagent Total volume * IMPORTANT: DO NOT use less than 60 µg (ten 15-cm plates) to package a batch of DECIPHER or 27K library. A smaller amount may cause shRNA insert representation to be adversely affected. 4. Add 900 µl of Lipofectamine Reagent to 12 ml of D-MEM medium without serum or antibiotics in order to make a convenient master mix. Mix gently. 10X plates 12,000 µl 900 µl 12,900 µl Component D-MEM, no FBS, no antibiotics Lipofectamine Total volume 5. Add the diluted Lipofectamine Reagent (from step 4) to the DNA / Plus Reagent complex (from step 3), mix gently by flicking the tube or vortexing and incubate at room temperature for 15 min. 6. Add 2.5 ml of the DNA / Plus Reagent / Lipofectamine Reagent complex (from step 5) to each 15cm plate from step 2, and mix complexes with medium by gentle rotation. Take care not to dislodge cells from the plate. Incubate at 37°C in the CO2 incubator for 24 hours. Day 2 – DNAse I Treatment 7. At 24 hours post-transfection, replace the medium containing complexes with fresh 30 ml D-MEM medium supplemented with 10% FBS, DNase I (1 U/ml), MgCl2 (5 mM), 20mM HEPES pH7.4. Continue incubation in the CO2 incubator at 37°C overnight. Overnight DNase I treatment before harvesting virus does not negatively affect lentiviral titer or infectivity and helps prevent undesirable carryover of plasmid library into the virus prep. [email protected] 7 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net NOTE: Failure to change the media the day after transfection results in large carryover of plasmid (free and/or Lipofectamine-bound) in your lentiviral prep. This may cause problems with most downstream molecular biology applications, especially whenever there is a PCR step involved. Day 3 – Collect Lentiviral Supernatant 8. At 48 hours post-transfection, collect all 30 ml of the virus-containing medium from each plate and filter the supernatant (300 ml) through a Nalgene 0.2 µm PES filter (a low protein binding filter) to remove debris and floating packaging cells. Failure to filter supernatant could result in carry-over of cells into your lentiviral prep. NOTE: Usually, the peak of virus production is Supernatant can also be collected again at 72 hours hour supernatant with 30 ml of fresh D-MEM medium pH7.4 and continue incubation in the CO2 incubator at achieved at 48 hours post-transfection. post-transfection—replace the collected 48supplemented with 10% FBS, 20mM HEPES 37°C for 24 hours. CAUTION: You are working with infectious lentiviral particles at this stage. Please follow the recommended guidelines for working with BSL-2 safety class materials (see Safety Guidelines). 9. Aliquot and store the supernatant at –80°C. lentiviral titer with each cycle. Freezing and thawing usually results in ~20% loss of Cellecta offers lentiviral packaging services. Please contact us at [email protected] or visit http://www.cellecta.com/products-and-services/lentiviral-packaging/ for more information. [email protected] 8 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net D. Transduction Protocols and Lentiviral Titer Estimation Transduction Lentiviral transductions are performed by mixing cells and virus in culture media supplemented with Polybrene®. For both adherent and suspension cells, transductions are initiated in suspension and carried out overnight. Adherent cells are allowed to adhere to substrate during transduction and are transduced at a cell density that allows for 2-3 population doublings before reaching confluence. Suspension cells are typically transduced at higher density than standard growth density, and then they are diluted to standard growth density 18-24 hours after transduction. Check Toxicity of Polybrene Polybrene is a polycation that neutralizes charge interactions to increase binding between the lentiviral envelope and the plasma membrane. The optimal concentration of Polybrene depends on cell type and may need to be empirically determined. Excessive exposure to Polybrene can be toxic to some cells. Before conducting the titer estimation experiment, we recommended performing a Polybrene toxicity titration in target cells. Grow cells in complete culture medium with a range of Polybrene concentrations (0 µg/ml, 1 µg/ml, 2 µg/ml, 3 µg/ml, 4 µg/ml, 5 µg/ml) for 24 hours, and then replace old medium with Polybrene-free complete culture medium. Grow cells for an additional 72 hours, and then check toxicity by counting viable cells. For your experiments, use the highest concentration of Polybrene that results in less than 10% cell toxicity compared to no Polybrene (typically, 5 µg/ml is recommended). For some cell types, you cannot use Polybrene. Protocol For Titering lentiviral stock (RFP assay) Lentiviral shRNA vectors that express the fluorescent protein TagRFP (excitation ~560nm emission ~590nm) allow lentiviral titer estimation by flow cytometry (RFP assay) or by a combined flow cytometry/puromycin resistance assay (RFP/PuroR assay). To check lentiviral titer, we recommend always using the same cells you will use in the screen. Most of the commonly used mammalian cell lines can be effectively transduced by lentiviral constructs. Relative titers can vary up to 50-fold depending on the chosen cell line. Transduction (HEK293 cells): The following protocol has been optimized for HEK293 cells. For other adherent cell types, parameters such as media, growth surface, time of detection, etc. will have to be adjusted. Day 1 1. Quickly thaw the lentiviral particles in a water bath at 37°C. Transfer the thawed particles to a laminar flow hood, gently mix by rotation, inversion, or gentle vortexing, and keep on ice. CAUTION: Only open the tube containing the lentiviral particles in the laminar flow hood. NOTE: Unused lentiviral stock may be refrozen at –80°C, but it will typically result in a loss of about ~20% in titer. 2. Trypsinize and resuspend HEK293 cells to a density of 1 × 105 cells/ml in D-MEM supplemented with 10% FBS and 5 µg/ml Polybrene. Aliquot 1 ml/well in a 12-well plate and add 0 µl, 3 µl, 10 µl, 33 µl, and 100 µl of lentiviral stock (supernatant filtered to remove cells and cell debris, not concentrated) to six different wells. If concentrated virus is used, scale down virus volumes accordingly. Mix and return cells to CO2 incubator. Grow cells under standard conditions for 24 hours. NOTE: It is important to accurately record the original # of cells at Time of Transduction, as this is critical in titer calculation. For adherent cells other than HEK293, choose a different # of cells at time of transduction, depending on cell size. As a rule of thumb, cells should be transduced at such a density such that they would become confluent in ~48 hours. For example, for HeLa cells, the suggested cell # is 50,000 cells/well in a 12-well plate. [email protected] 9 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Day 2 3. Between 16h-24 hours post-transduction, replace media with fresh D-MEM supplemented with 10% FBS and without Polybrene. Return cells to CO2 incubator, and grow under standard conditions for additional 48 hours. Avoid confluence: trypsinize and re-plate cells if needed. Day 4 (72 hours after transduction) 4. Detach cells from the plate by trypsin treatment, block trypsin with FBS/media, centrifuge, resuspend in 1X D-PBS, and determine the % of transduced (RFP-positive) cells by flow cytometry. NOTE: Attempting to determine the % of transduced cells by fluorescence microscopy is NOT RECOMMENDED. IMPORTANT: Flow cytometry settings to detect RFP-positive cells are the following: Excitation: 561nm (530nm laser is still acceptable), Emission: 600/20 band-pass filter, or similar (for TagRFP). 5. Proceed to Lentiviral Titer estimation (RFP assay). Alternative Transduction protocol (spinoculation) for hard to transduce cells The following protocol has been optimized for K-562 cells. For other cell types, parameters such as media, growth surface, time of detection, etc. will have to be adjusted. 1. K-562 cells are transduced (“infected”) using spinoculation. This is performed using multi-well tissue culture plates and a tabletop centrifuge capable of 1,200 × g and centrifugation of multiwell plates. 2. Grow K-562 cells and maintain them between 2 × 105 and 1 × 106 cells/ml. Do not let them become too dense or let the medium become yellow at any point. 3. For lentiviral library titration, K-562 cells are resuspended at 2 × 106 cells per ml in RPMI 10%FBS supplemented with 20mM HEPES pH7.4 and Polybrene 5 µg/ml. 0.5-ml aliquots are placed into each well in a 24-well plate (1 × 106 cells/well total). This cell density has proven effective for many suspension cell lines in-house at Cellecta. To each cell-containing well, add increasing amounts of lentiviral stock to be titered. For 100-fold concentrated lentiviral stock, for example, add 0 µl, 0.3 µl 1 µl, 3 µl, and 10 µl virus. Close the plate, mix by gentle agitation, wrap the perimeter with parafilm, and place the plate into centrifuge with an appropriate balance and centrifuge at 1,200 × g at +25°C for 2 hours. 4. Following centrifugation, remove plate(s) from centrifuge, carefully remove parafilm, and place in incubator. After 3 hours, “feed” cells with 0.5 ml additional complete medium per well (no Polybrene). 5. 24 hours after spinoculation, resuspend cells at 2 × 105 cells/ml in RPMI 10% FBS in the appropriate culture vessel and grow for additional 48 hours. 6. 72 hours after spinoculation, perform titer as previously described. NOTE: Use larger vessels for large-scale genetic screen transductions. Scale up all volumes accordingly. [email protected] 10 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Lentiviral Titer estimation (RFP assay) Lentiviral titer is measured as Transduction Units/ml (TU/ml). One TU produces one integration event in target cells. Integration events can be calculated from observed % of transduced cells according to the table below. TITER CHART 100 90 80 % infected cells 70 60 50 40 30 20 10 0 0 0.5 1 1.5 2 2.5 MOI (integrations/cell) The % of infected cells is determined by flow cytometry (excitation=561nm emission=600/20 for TagRFP) by observing the % of RFP+ cells in the transduced cell sample. When the % of infected cells is at or below 20%, the # of integrations is (with good approximation) equivalent to the # of transduced cells. At higher transduction efficiencies, the fraction of transduced cells bearing multiple integrations becomes higher and higher, so that the increase in % of transduced cells relative to integration events/cell is no longer linear. Using the table below, MOI (MOI=multiplicity of infection = integrations/cell) can be calculated with good accuracy in the range 0.2-1.5 MOI. Titer is calculated according to the TITER FORMULA below: TU/ml = (# of cells at Transduction) x MOI / (ml of Viral Stock used at Transduction) Example: IF: The original # of cells at Transduction was 100,000, and The volume of virus stock used was 10 µl, and The observed % of transduced (RFP+) cells is 25%, THEN: The calculated MOI is 0.3, and The TITER is: 100,000 × 0.3 / 0.01 = 3,000,000 TU/ml Once titer is estimated, the amount of Lentiviral Stock necessary to transduce any given # of target cells at any transduction efficiency (range of 10-80% infected cells) can be backward calculated from the TITER FORMULA and TITER CHART above. [email protected] 11 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Example: To transduce 20,000,000 cells at 50% transduction efficiency, with a Lentiviral Stock titer of 3,000,000 TU/ml, we calculated the required amount of Lentiviral Stock as follows: 1. We calculate the required MOI to achieve 50% transduction efficiency, using the TITER CHART: 50% transduction efficiency = 0.7 MOI 2. We calculate the volume of Lentiviral Stock required using the TITER FORMULA: TU/ml = (# of cells at Transduction) x MOI / (ml of Viral Stock used at Transduction) 3,000,000 = 20,000,000 × 0.7 / (ml Viral Stock) Viral Stock = 20,000,000 × 0.7 / 3,000,000 = 4.67 ml E. Protocols for Genetic Screens with Pooled shRNA Libraries To ensure reproducible and reliable results when using pooled shRNA libraries, it is critical that you transduce enough cells to maintain sufficient representation of each shRNA construct present in the cellular library. The number of cells stably transduced with the shRNA library at the time of transduction should exceed the complexity of the shRNA library by at least 200-fold or optimally 1,000-fold. After transduction, it would be best never to discard any cells at any time during the experiment (e.g. at splitting steps). If the number of cells is too high to grow and you are forced to discard a fraction of the cells (as it is often the case in negative selection/drop-out screens), the number of remaining cells should always exceed the complexity of the library by at least 1,000-fold (e.g. keep at least 2.7 × 107 cells after every splitting step, for a 27K library). IMPORTANT: Before discarding any cells at splitting step, make sure you pool ALL cells from each same replicate together. Additionally, when using pooled shRNA libraries, you should consider that the higher the percentage of transduced cells and MOI, the higher the percentage of infected cells that will bear two or more different shRNA constructs. For most RNAi screens, we recommend optimizing transduction conditions and performing genetic screen transductions at no more than 0.5 MOI (40% transduction efficiency). NOTE: We highly recommend titering the library directly in your target cells prior to beginning your experiment. Once conditions are established to achieve ~40% transduction efficiency in the titering assay, accurately scale up all conditions to accommodate the larger amount of transduced cells needed for the genetic screen. EXAMPLE: Translating RFP-based titer into PuromycinR-Titer (RFP/PuroR assay) If puromycin selection of transduced cells is going to be performed in the screen (as it is recommended in most negative selection/drop-out screens), it is important to determine beforehand the minimum amount of puromycin required to kill untransduced cells. This can be done empirically by generating a kill curve, as follows: Puromycin Kill Curve. Aliquot cells in a 12-well plate at such a density so they are at 72 hours from confluency. Add puromycin at 0 µg/ml, 0.5 µg/ml, 1 µg/ml, 2 µg/ml, 5 µg/ml, and 10 µg/ml in six different wells. Mix and return cells to incubator. Grow cells under standard conditions for 42-72 hours. For puromycin, use the lowest concentration that kills >90% of cells in 42-72 hours. PuromycinR-Titer. If puromycin selection of transduced cells is going to be performed in the screen, the fraction of RFP+ cells (at a given MOI) that will survive puromycin selection must calculated beforehand. Even though RFP and Puro-resistance markers are expressed from the same promoter, not all cells expressing detectable RFP are guaranteed to be puro-resistant. A threshold of PuroR [email protected] 12 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net expression is required to confer puromycin resistance. Depending on cell type, such a threshold is associated with different levels of RFP co-expression. Depending on the MOI used, a different % of RFP+ cells will express enough PuroR to survive puromycin selection (i.e. the higher the MOI, the higher the % of multiple integrants, so the higher the % of RFP+ cells expressing higher levels of puroR). In order to calculate which fraction of RFP+ cells are going to survive puromycin selection, the following procedure is strongly suggested: 1. Titer virus in target cell line, by flow cytometry (see Lentiviral Titer estimation (RFP assay) in the Transduction Protocols and Lentiviral Titer Estimation Section). 2. Based on assessed titer, perform a small-scale transduction aiming at 50% infected cells: 3 days after transduction, split cells into 2 samples, grow cells +/- puromycin for an additional 3 days, then analyze both samples by flow cytometry. 3. By looking at the RFP intensity of puromycin-treated cells, calculate the % of cells that survived puromycin selection. The figure below shows FACS analysis of transduced cells—no puromycin selection (blue), puromycin selection (red). 50% of cells were RFP+, 24% of the RFP+ cells were also puromycin-resistant (12% of total). IMPORTANT: The % of RFP+ cells that are also puromycin-resistant is dependent on MOI, as it increases with the increase of % of RFP+ cells bearing multiple integrations. In the example above, 24% of RFP+ cells (12% of total) were puromycin-resistant when cells were infected at MOI 0.7 (50% RFP+ cells). If the same cells would be infected at the recommended MOI of 0.5 (40% RFP+ cells), less than 24% of RFP+ cells would also be puromycin-resistant cells. Conversely, if cells would be infected at MOI 2 (85% RFP+ cells), a much higher % than 24% of RFP+ cells would also be puromycin-resistant, due to high % of RFP+ cells bearing multiple integrants and therefore expressing high levels of the puromycin-resistance gene. In the case described above, a 27K library genetic screen started with at least 46 × 106 cells per replicate and transduction. Cells were infected at MOI 0.7 (50% transduction efficiency) to obtain 23 × 106 infected (RFP+) cells, of which about 5.5 × 106 will be puroresistant (200 puro resistant cells/shRNA). In your screening experiment, however, we do not recommend using an MOI greater than 0.5. IMPORTANT: Using higher MOIs to achieve >40% RFP+ cells in order to obtain ~20% or more puro-resistant cells is not recommended. It is advised to limit the RFP-based MOI to 0.5 (40% RFP+ cells) and use enough cells at transduction to obtain the desired amount of puromycin-resistant transduced cells (at least 200 cells/shRNA). IMPORTANT: When performing lentiviral transductions for a genetic screen, make sure to use exactly the same conditions as in library titering. Accurately scale up volumes, surfaces, cell number, and reagents to be used. [email protected] 13 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Example of a Negative Selection Viability “Drop out” RNAi Screen: Identification of genes essential for viability of AR-1 negative Human Prostate Cancer DU145 cells (treatment is time). The DECIPHER Human Module 1 pooled lentiviral shRNA library of 27K complexity was used. Six independent transductions were performed. Each transduction consisted of 2 × 107 cells infected at 30% efficiency based on RFP/PuroR Assay (~6 × 106 infected cells). Each transduction was treated as an independent sample. Each independent sample had an estimated average of ~200 clones per shRNA. NOTE: In most cases, 40% transduction efficiency is recommended. Day 1. Cells were trypsinized and resuspended to a density of 2 × 105 cells/ml in D-MEM supplemented with 10% FBS and 5 µg/ml Polybrene. 25 ml of cells were aliquoted to each 15-cm plate (4 plates per replicate, 2 × 107 cells per replicate), and enough virus was added to achieve ~1.5 × 106 infected cells per plate (~6 × 106 cells per replicate). Cells were returned to CO2 incubator and grown under standard conditions for 24 hours. Day 2. At 18 hours post-transduction, three (3) samples were harvested and stored as frozen cell pellets (baseline samples). To the three (3) remaining samples, media containing virus/Polybrene was replaced with fresh media (without Polybrene). Day 3. At 72 hours post-transduction, puromycin was added to the samples at a final concentration of 1 µg/ml (about 90% RFP+ cells were expected to be also puromycin resistant, based on RFP/PuroR titer assay). Puromycin selection was carried out for 48 hours. NOTE: In most cases, it is recommended to wait 72 hours after transduction before puromycin selection. NOTE: Keeping cells under puromycin selection for more than 48 hours is not required. Day 9. Samples were harvested and stored as frozen cell pellets. Genomic DNA was then extracted and purified from treated and untreated samples. barcodes were amplified from genomic DNA and enumerated by HT sequencing. shRNA insert Cellecta offers complete RNAi screening services. Please contact us at [email protected] or visit http://www.cellecta.com/products-and-services/RNAi-screening-services/ for more information. [email protected] 14 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net F. Genomic DNA Extraction for Barcode Amplification and HT Sequencing Identification of shRNA barcodes in the experimental samples requires amplification of the barcode portion of the integrated lentiviral constructs from sample genomic DNA. Subsequent high-throughput sequencing of barcodes by the Illumina GAIIx or HiSeq is done to quantify each barcode and generate digital expression data using Deconvolution software. We currently do not support HT sequencing of samples on the Illumina MiSeq. Cellecta now offers sample prep, HT sequencing, and analysis services. Please contact us at [email protected] or visit http://www.cellecta.com/products-and-services/pooled-lentivirallibraries/HT-sequencing-and-analysis/ for more information. Recommended Protocol NOTE: Use of disposable tubes is highly recommended in order to avoid contamination. 1. Suspend cell pellet in 5 ml QIAGEN buffer P1 (with RNaseA) in 15 ml POLYPROPYLENE (phenol/chloroform resistant), BD FALCON screw-cap centrifuge tube (12,000 RCF rated, BD Biosciences Cat.# 352196). 2. Add 0.25 ml 10%SDS, mix and incubate 5 minutes at RT. 3. Using an ultrasonic homogenizer (see Required Materials), sonicate to shear DNA into 10-100 kb sized fragments. To prevent cross-contamination, thoroughly wash the ultrasound head with running water and dry-up with clean paper towel between samples. 4. Add 5 ml phenol/chloroform pH8.0 solution, vortex hard and spin down 60 min, +20°C at 8,000 rpm in JA-14 or equivalent rotor (Beckman). 5. You should have about 5 ml of clear upper phase. Transfer 4 ml of upper phase to new 15 ml DISPOSABLE screw cap tube (same as in Step 1). 6. Add 0.5 ml 3M Sodium Acetate, 4 ml isopropanol, mix well, and spin down 30 min, +20°C at 8,000 rpm in JA-14 or equivalent rotor. 7. In order to have a more visible pellet, compacted at the bottom of the tube, it is recommended to incubate overnight at RT before centrifugation. IMPORTANT: If starting material is less than 5 million cells, add carrier before centrifugation (linear polyacrylamide, 25 µg/ml final) and spin down for a longer time (60 min). 8. Discard supernatant, add 10 ml 70% ethanol, spin down 5 min, +20°C at 8,000 rpm in JA-14 or equivalent rotor. 9. Discard supernatant and air-dry pellet. 10. Dissolve DNA pellet in appropriate volume of dH2O to a concentration of approximately 2 mg/ml. Expected yield is about 10 µg per 1 million cells. 11. Incubate 30 minutes at +80°C before spectrophotometer reading. [email protected] 15 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net G. Amplification of shRNA-specific Barcodes from Genomic DNA The pooled barcodes should be amplified from 200 µg of genomic DNA (isolated from cell samples in the previous step) by two rounds of PCR using Titanium Taq DNA polymerase mix (Clontech-Takara, see Required Materials). Use the entire amount of genomic DNA and a proportionally fewer number of 100-µl reactions per sample when amplifying barcodes from samples generated by positive selection screens. The protocol was optimized using an ABI GeneAmp PCR System 9700. Use of other PCR enzymes and/or thermal cyclers may require additional optimization. The lentiviral shRNA library and PCR primer designs include sequences complementary to the sequences of the immobilized primers necessary for generating amplification clusters in Illumina’s GAIIx or HiSeq Flow Cells. Our library design is only compatible with Single-Read Flow Cells (in the Single-Read Cluster Generation Kit), because our primers are not complementary to the sequences immobilized on Paired-End flow cells (in the Paired-End Cluster Generation Kit). See Required Materials for the appropriate Illumina catalog numbers. HT sequencing of samples on the Illumina MiSeq is not supported. Please use 10 ng of plasmid shRNA library as an amplification control in the first round of PCR, and use the subsequent PCR products in all remaining steps. First Round of PCR 1. For each sample, prepare 4 × 100 µl reactions containing a total of 200 µg of genomic DNA: ___ 3 3 2 10 ___ 1 100 µl µl µl µl µl µl µl µl 94°C, 94°C, 65°C, 72°C, 68°C, Genomic DNA (50 µg) Forward 1st round PCR primer* (10 µM) Reverse 1st round PCR primer* (10 µM) 50X dNTP Mix (10 mM each) 10X Titanium Taq Buffer Deionized water 50X Titanium Taq Total volume 3 minutes 30 seconds 10 seconds 20 seconds 2 min 1 cycle 16 cycles 1 cycle * Please see Appendix for primer sequences for vectors with HTS4 and HTS6 shRNA cassettes. For other vectors, please contact Cellecta. [email protected] 16 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Second Round of PCR The second round of PCR—nested PCR—is required in order to significantly reduce genomic DNA carryover into the samples used for HT sequencing. Amplify each DNA sample with the Forward and Reverse 1st round primer set* and perform HT sequencing on one sample per lane (in the flow cell) with the GexSeq* primer. 1. Combine together the 4 × 100 µl First Round PCR reactions and use a 2 µl aliquot in the second round of analytical PCR with nested primers in each 100 µl reaction: 2 5 5 2 10 75 1 100 µl µl µl µl µl µl µl µl 94°C, 94°C, 65°C, 72°C, 68°C, First Round PCR Product Forward 2nd round PCR primer* (10 µM) Reverse 2nd round PCR primer* (10 µM) 50X dNTP Mix (10 mM each) 10X Titanium Taq Buffer Deionized water 50X Titanium Taq Total volume 3 minutes 30 seconds 10 seconds 10 seconds 2 min 1 cycle 10, 12, or 14 cycles 1 cycle * Please see Appendix for primer sequences for vectors with HTS4 and HTS6 shRNA cassettes. For other vectors, please contact Cellecta. NOTE: Avoid overcycling of PCR reactions—this will usually result in the generation of a longer fragment that corresponds to a fusion double barcode product. The amplified pooled barcode cassettes are then analyzed on a 3.5% agarose-1XTAE gel (load 5 µl/lane). The results should reveal a bright band of amplified barcode products (HTS3 cassette: 106bp; HTS4: 255-bp; HTS6: 251-bp). The goal of this analytical PCR step is to optimize the starting amount of First Round PCR product and the number of cycles (if necessary) in order to achieve equal intensities of a single band across all DNA samples from the genetic screen. Repeat second-round amplification of barcodes from each sample using the optimized volume of First Round PCR product, 2 × 100 µl of Second Round PCR product per sample, and 12-14 cycles of PCR. Set up 2 × 100 µl reactions for each sample containing an adjusted “equal” amount of First Round PCR product (2 µl or more): 2 5 5 2 10 75 1 100 µl µl µl µl µl µl µl µl 94°C, 3 94°C, 65°C, 72°C, 68°C, 2 [email protected] First Round PCR Product Forward 2nd round PCR primer (10 µM) Reverse 2nd round PCR primer (10 µM) 50X dNTP (10 mM each) 10X Titanium Taq Buffer Deionized water 50X Titanium Taq Total volume minutes 30 seconds 10 seconds 10 seconds min 1 cycle 12 or 14 cycles 1 cycle 17 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Analyze the PCR products by gel-electrophoresis on a 3.5% agarose-1XTAE gel in order to ensure equal yields of amplified barcodes for all samples. Combine amplified barcodes from the 2 × 100 µl Second Round PCR reactions and purify the samples as follows: 1. Purify the PCR product with the QIAquick PCR purification kit (QIAGEN) following the manufacturer’s protocol, 2. Separate by electrophoresis in a preparative 3.5% agarose-1XTAE gel, 3. Cut out band and extract DNA from the gel using the QIAquick gel purification kit (QIAGEN), and 4. Quantitate using A260 nm measurement using NanoDrop spectrophotometer (or equivalent) and adjust concentration to 10nM (e.g. 0.75 ng/µl for 106-bp product (HTS3), or ~1.8 ng/µl for 255bp (HTS4) or 251-bp (HTS6) product). H. HT Sequencing of Pooled shRNA-specific Barcodes on Illumina’s GAIIx or HiSeq HT sequencing of pooled amplified barcodes can be performed on the Illumina GAIIx (~20-30 million reads per sample) or HiSeq (~80-100 million reads per sample) using the GexSeq* sequencing primer and following the manufacturer’s protocol. The final concentration of GexSeq* primer in the reaction should be 500 nM. For the cluster generation step, use 20 fmoles (2 µl of 10 nM PCR product) of the gel-purified band from the 2nd round of PCR. The number of cycles (read length) required depends on the length of the barcode and whether there is an additional sequence, e.g. clonal barcode (contact Cellecta for details). Required read lengths for different vectors are indicated below: The shRNA library and PCR primer designs include sequences complementary to the sequences of the immobilized primers necessary for generating amplification clusters in Illumina’s GAIIx or HiSeq flow cells. Our design is only compatible with Single-Read Flow Cells (in the Single-Read Cluster Generation Kit), because our primers are not complementary to the sequences immobilized on Paired-End flow cells (in the Paired-End Cluster Generation Kit). Protocol for HT sequencing on Illumina HiSeq platform. 1. 2. 3. 4. Adjust purified PCR samples to 10nM (1.7 ng/µl) concentration. For cluster generation step – use Illumina Single-Read (SR) flow cell, and for each lane add 2 µl of each sample and add PhiX174 control template based on standard Illumina protocol. For HT sequencing step – Add GexSeq* primer (10 µM, i.e. 20x) to the PhiX174 primer to a final concentration of 0.5 µM. Run HT sequencing reaction for the appropriate number of cycles with GexSeq/PhiX174 primer mix. See Required Materials for a list of recommended Illumina kits for HT Sequencing of samples transduced with a Cellecta library. * Please see Appendix for HT sequencing primer sequences for vectors with HTS3, HTS4, and HTS6 shRNA cassettes. For other vectors, refer to the Product Analysis Certificate that came with the product or contact Cellecta. Cellecta now offers sample prep, HT sequencing, and analysis services. Please contact us at [email protected] or visit http://www.cellecta.com/products-and-services/pooled-lentivirallibraries/HT-sequencing-and-analysis/ for more information. I. Barcode Enumeration (Conversion of raw sequencing data to number of reads for each barcode) For DECIPHER shRNA Libraries, step-by-step protocols for barcode deconvolution and enumeration are included with the downloadable software available on the DECIPHER Project website at http://www.decipherproject.net/software/. For Custom shRNA Libraries, please contact Cellecta Technical Support at [email protected]. [email protected] 18 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net J. Troubleshooting Low Lentiviral Titer (<106 TU/ml in supernatant) 1. Poor transfection efficiency (48 hour post-transfection, less than 80% of 293T cells are very brightly fluorescent) Problem: 293T Cells have too high or too low density Solution: Plate fewer or more cells in order to have about 80% confluency at time of transfection. Problem: Plasmid DNA/Lipofectamine/Plus Reagent ratios are incorrect Solution: Optimize the ratios using the guidelines provided in the Lipofectamine protocol. 2. Inefficient production of the virus Problem: 293T Cells are of poor quality Solutions: • Optimize growth conditions, check growth medium, and don’t grow 293T cells for more than 20 passages. • Check for mycoplasma contamination. • Do not overgrow the cells (do not allow the cells to reach more than 90% confluency in order to keep the culture continuously in logarithmic growth phase). Problem: Lentiviral supernatant harvested too early or too late Solution: Harvest supernatant 48 hours and 72 hours after transfection. Problem: 293T cell media is too acidic at time of virus harvesting Solution: Make sure to replace media 24 hours before harvesting, and make sure to supplement media with HEPES pH 7.4 20mM final. 3. Inefficient transduction of titering cells See below. Inefficient Transduction of Packaged shRNA Library 1. Poor transduction efficiency Problem: Target cells have too high or too low density Solution: Plate fewer or more cells in order to have 20-50% confluency at transduction stage. Problem: Target cell line may be difficult to transduce Solutions: • Use a higher concentration of lentiviral particles. • Perform “Spinoculation” to improve transduction efficiency. • Check to see if Polybrene was added at 5 µg/ml. Problem: Wrong amount of Polybrene added during transduction stage Solution: If Polybrene is toxic to the target cells, optimize Polybrene concentration in the range of 0 – 5 µg/ml by performing a toxicity titration as described in the Transduction Protocols and Lentiviral Titer Estimation Section. Problem: Loss of lentiviral titer during storage Solution: Ensure storage of aliquoted packaged shRNA library at –80°C. Each freeze-thaw cycle typically causes reduction of the titer by ~20%. Use a fresh stock for transduction. Problem: The RFP assay is performed too early [email protected] 19 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Solution: Normally, the maximal expression of RFP from the integrated provirus is expected to develop by 72 hours after transduction. However, some cells exhibit delayed expression. Try the assay at a later time, such as 96 hours. Problem: The RFP assay is performed with the wrong flow cytometry settings. Solution: RFP+ cells are to be detected using a 561nm laser for excitation (530nm still acceptable) and 600/20 band-pass filters (or similar) for detection (for TagRFP). Using blue laser (488nm) for excitation leads to gross underestimation of viral titer. Problem: In the RFP assay, the % of transduced cells is determined by fluorescence microscopy instead of flow cytometry. Solution: Use flow cytometry. 2. Transduction affects target cell viability Problem: Polybrene is toxic for target cells Solution: Optimize the concentration and exposure time to Polybrene during the transduction step. For some sensitive cells, Polybrene should not be used. Problem: Virus-containing conditioned media is toxic to target cells. Solution: Concentrate and resuspend the virus in target cell growth media, PBS 10% FBS, or PBS 1% BSA. 3. No expression of RFP or PuroR (or shRNAs) in target cells Problem: The UbiC (or U6 shRNA) promoter is not functional in target cells. Solutions: • Change the target cells. • Contact Cellecta at [email protected] to have the library cloned in another vector with different promoter. Difficulties with Probe Preparation and HT Sequencing 1. No PCR Product Problem: Incorrect primers or bad reagents used, or missing reagents. Solutions: • Include 10 ng of plasmid library DNA as a positive control. If it produces the correct amplification product, the problem lies with the genomic DNA or previous PCR prep. If not, confirm use of the correct primers and reagents. • Verify that primer sequences are correct. For vectors with HTS3, HTS4, or HTS6 shRNA cassettes, please see Appendix. For all other vectors, please contact Cellecta at [email protected]. 2. No barcodes present in HT Sequencing results Problem: Incorrect primer used in Illumina-Solexa Cluster Generation step. Solution: Ensure that you or the HT Sequencing core facility uses the proper GexSeq Sequencing primer (see Appendix), NOT the Sequencing primer that comes with the Illumina Cluster Generation Kit. Problem: Incorrect Cluster Generation kit used. Solution: Ensure that you or the HT Sequencing core facility uses the proper Single-Read Cluster Generation Kit (see Required Materials). [email protected] 20 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net L. Technical Support DECIPHER Project Users: For help with using DECIPHER Pooled Lentiviral shRNA Libraries, please email technical support at [email protected] with the answers to the questions below (if applicable). Library Used: 1. 2. Which library did you use, and which Module(s)? What are the lot numbers? Packaging the Library: 3. 4. What was the lentiviral titer, and what was the total number of TU packaged? How was the virus concentrated? (if applicable) Transducing Target Cells: 5. 6. 7. 8. 9. What MOI did you use to transduce your target cells? What target cells did you use? How many replicates did you use? (i.e. duplicate, triplicate, etc.) Did you use puromycin after transduction, and at what concentration? For how long did you use puromycin on the cells? RNAi Screen: 10. Could you briefly explain your experiment? 11. How many infected cells were used? Sample Preparation & HT Sequencing 12. 13. 14. 15. What HT sequencing system and which Illumina HT Sequencing Kits did you use? How much PCR product was used for HT Sequencing? How many sequences were read per sample? Would you be able to send us the raw data so that it may help us diagnose the issue? All Users: For additional information or technical assistance, please refer to the questions above and contact us by phone or email: Phone: Toll-Free: Fax: +1 (650) 938-3910 +1 (877) 938-3910 +1 (650) 938-3911 E-mail: Technical Support: General Information: Sales: Orders: Blog: [email protected] [email protected] [email protected] [email protected] http://www.cellecta.com/blog/ Postal Mail: Cellecta, Inc. 320 Logue Ave. Mountain View, CA 94043 For more information about Cellecta’s products and services, please visit our web site at http://www.cellecta.com. [email protected] 21 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net M. Safety Guidelines The HIV-based lentivector system is designed to maximize its biosafety features, which include: • • • • • A deletion in the enhancer of the U3 region of 3’ΔLTR ensures self-inactivation of the lentiviral construct after transduction and integration into genomic DNA of the target cells. The RSV promoter upstream of 5’LTR in the lentivector allows efficient Tat-independent production of lentiviral RNA, reducing the number of genes from HIV-1 that are used in this system. Number of lentiviral genes necessary for packaging, replication and transduction is reduced to three (gag, pol, rev). The corresponding proteins are expressed from different plasmids lacking packaging signals and share no significant homology to any of the expression lentivectors, pVSV-G expression vector, or any other vector to prevent generation of recombinant replication-competent virus. None of the HIV-1 genes (gag, pol, rev) will be present in the packaged lentiviral genome, as they are expressed from packaging plasmids lacking packaging signal—therefore, the lentiviral particles generated are replication-incompetent. Lentiviral particles will carry only a copy of your expression construct. Despite the above safety features, use of HIV-based vectors falls within NIH Biosafety Level 2 criteria due to the potential biohazard risk of possible recombination with endogenous lentiviral sequences to form self-replicating virus or the possibility of insertional mutagenesis. For a description of laboratory biosafety level criteria, consult the Centers for Disease Control Office of Health and Safety Web site at: http://www.cdc.gov/biosafety/publications/bmbl5/bmbl5_sect_iv.pdf It is also important to check with the health and safety guidelines at your institution regarding the use of lentiviruses and follow standard microbiological practices, which include: • • • • • Wear gloves and lab coat at all times when conducting the procedure. Always work with lentiviral particles in a Class II laminar flow hood. All procedures are performed carefully to minimize the creation of splashes or aerosols. Work surfaces are decontaminated at least once a day and after any spill of viable material. All cultures, stocks, and other regulated wastes are decontaminated before disposal by an approved decontamination method such as autoclaving. Materials to be decontaminated outside of the immediate laboratory area are to be placed in a durable, leakproof, properly marked (biohazard, infectious waste) container and sealed for transportation from the laboratory. [email protected] 22 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net N. References Cellecta Research and Collaborations Khorashad JS, Mason CC, Kraft IL, Reynolds KR, Pomicter AD, Eiring AM, Zabriskie MS, Iovino AJ, Heaton W, Tantravahi SK, Kauffman M, Schacham S, Chenchik A, Bonneau K, O'Hare T, Deininger MW. (2013) An Unbiased shRNA Library Screen Identifies Nucleocytoplasmic Transport As a Potential Target For Treatment Of Chronic Myeloid Leukemia. Blood. 122(21):2707. Nolan-Stevaux O, Tedesco D, Ragan S, Makhanov M, Chenchik A, Ruefli-Brasse A, Quon K, Kassner PD. (2013) "Measurement of Cancer Cell Growth Heterogeneity through Lentiviral Barcoding Identifies Clonal Dominance as a Characteristic of In Vivo Tumor Engraftment." PLoS One. Jun 26;8(6):e67316. Print 2013. PMID: 23840661 Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, Gudkov AV. (2013) "p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs." Proc Natl Acad Sci U S A. Jan 2;110(1):E89-98. doi:10.1073/pnas.1216922110. Epub 2012 Dec 10. PMID: 23236145 Neznanov N, Komarov AP, Neznanova L, Stanhope-Baker P, Gudkov AV. (2011) "Proteotoxic stress targeted therapy (PSTT): induction of protein misfolding enhances the antitumor effect of the proteasome inhibitor bortezomib." Oncotarget. Mar;2(3):209-21. PubMed PMID: 21444945; PubMed Central PMCID: PMC3260823. Singhal R, Deng X, Chenchik AA, Kandel ES. (2011) "Long-distance effects of insertional mutagenesis." PLoS One. Jan 5;6(1):e15832. doi: 10.1371/journal.pone.0015832. PMID: 21246045 Tsujii H, Eguchi Y, Chenchik A, Mizutani T, Yamada K, Tsujimoto Y. (2010) "Screening of cell death genes with a mammalian genome-wide RNAi library." J Biochem. Aug;148(2):157-70. doi: 10.1093/jb/mvq042. Epub 2010 Apr 26. PubMed PMID: 20421362. Neznanov N, Gorbachev AV, Neznanova L, Komarov AP, Gurova KV, Gasparian AV, Banerjee AK, Almasan A, Fairchild RL, Gudkov AV. (2009) "Anti-malaria drug blocks proteotoxic stress response: anti-cancer implications." Cell Cycle. Dec;8(23):3960-70. Epub 2009 Dec 25. PubMed PMID: 19901558; PubMed Central PMCID: PMC2923591. Citations of Cellecta Products Kitambi SS, Toledo EM, Usoskin D, Wee S. (2014) Vulnerability of Glioblastoma Cells to Catastrophic Vacuolization and Death Induced by a Small Molecule. Cell 157:1-16. Boettcher M, Lawson A, Ladenburger V, Fredebohm J, Wolf J, Hoheisel JD, Frezza C, Shlomi T. (2014) High throughput synthetic lethality screen reveals a tumorigenic role of adenylate cyclase in fumarate hydratasedeficient cancer cells. BMC Genomics. 15(1):158. PMID: 24568598 Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, Bagdasarian L, Huber J, Lindeman A, Chen D, Romero R, Ramadan N, Phadke T, Haas K, Jaskelioff M, Wilson BG, Meyer MJ, Saenz-Vash V, Zhai H, Myer VE, Porter JA, Keen N, McLaughlin ME, Mickanin C, Roberts CW, Stegmeier F, Jagani Z. (2014) Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. PNAS. PMID: 24520176 Wolf J, Müller-Decker K, Flechtenmacher C, Zhang F, Shahmoradgoli M, Mills GB, Hoheisel JD, Boettcher M. An in vivo RNAi screen identifies SALL1 as a tumor suppressor in human breast cancer with a role in CDH1 regulation. Oncogene. 2013 Dec 2. doi: 10.1038/onc.2013.515. [Epub ahead of print] PubMed PMID: 24292671. Miyazaki H, Higashimoto K, Yada Y, Endo TA, Sharif J, Komori T, Matsuda M, Koseki Y, Nakayama M, Soejima H, Handa H, Koseki H, Hirose S, Nishioka K. Ash1l methylates lys36 of histone h3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013 Nov;9(11):e1003897. doi: 10.1371/journal.pgen.1003897. Epub 2013 Nov 7. PubMed PMID: 24244179; PubMed Central PMCID: PMC3820749. Wolf J, Dewi D, Fredebohm JA, Muller-Decker K, Flechtenmacher CA, Hoheisel J, Boettcher, M. (2013) "A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype." Breast Cancer Research 15:R109 doi:10.1186/bcr3576. Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM. (2013). "BET bromodomain inhibition suppresses TH17-mediated pathology." J Exp Med. Oct 21;210(11):2181-90. doi: 10.1084/jem.20130376. Epub 2013 Oct 7. PubMed PMID: 24101376; PubMed Central PMCID: PMC3804955. [email protected] 23 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Fredebohm J, Wolf J, Hoheisel JD, Boettcher M. (2013) "Depletion of RAD17 sensitizes pancreatic cancer cells to gemcitabine." J Cell Sci. Aug 1;126(Pt 15):3380-9. doi: 10.1242/jcs.124768. Epub 2013 May 17. PubMed PMID: 23687379. Review articles discussing Cellecta technology Diehl P, Tedesco D, Chenchik A. (2014) Use of RNAi screens to uncover resistance mechanisms in cancer cells and identify synthetic lethal interactions. Drug Discovery Today: Technologies. Available online 7 January 2014, ISSN 1740-6749, http://dx.doi.org/10.1016/j.ddtec.2013.12.002. Ward, T. M., Jegg, A. M., & Iorns, E. (2013). Next-Generation Sequencing for High-Throughput RNA Interference Screens. In Next Generation Sequencing in Cancer Research. (pp. 287-299). Springer New York. MaryAnn Labant. Genetic Engineering & Biotechnology News. June 15, 2012, 32(12): 1-28. doi:10.1089/gen.32.12.10. Thalyana Smith-Vikos. Genetic Engineering & Biotechnology News. April 15, 2013, 33(8): 20-22, 24. doi:10.1089/gen.33.8.09. Zhu, Z., & Huangfu, D. (2013). Human pluripotent stem cells: an emerging model in developmental biology. Development. 140(4), 705-717. Mulcahy, L. A., & Carter, D. R. (2013). RNAi2013: RNAi at Oxford. Journal of RNAi and gene silencing: an international journal of RNA and gene targeting research, 9, 486. Demir, K., & Boutros, M. (2012). Cell Perturbation Screens for Target Identification by RNAi. In Bioinformatics and Drug Discovery. (pp. 1-13). Humana Press. Campeau, E., & Gobeil, S. (2011). RNA interference in mammals: behind the screen. Briefings in functional genomics. 10(4), 215-226. Cellecta First-Generation shRNA Library References: Tsujii H, Eguchi Y, Chenchik A, Mizutani T, Yamada K, Tsujimoto Y (2010). “Screening of Cell Death Genes With a Mammalian Genome-wide RNAi Library.“ J Biochem. Apr 26. [Epub ahead of print] PubMed PMID: 20421362. Ossovskaya, V. S., G. Dolganov, et al. (2009). “Loss of function genetic screens reveal MTGR1 as an intracellular repressor of beta1 integrin-dependent neurite outgrowth.“ J Neurosci Methods 177(2): 322-33. Gumireddy, K., A. Li, et al. (2009). “KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer.“ Nat Cell Biol 11(11): 1297-304. Chen, Y., R. Cairns, et al. (2009). “Oxygen consumption can regulate the growth of tumors, a new perspective on the warburg effect.“ PLoS One 4(9): e7033. Xu, M., M. Takanashi, et al. (2009). “USP15 plays an essential role for caspase-3 activation during Paclitaxelinduced apoptosis.“ Biochem Biophys Res Commun 388(2): 366-71. Yeung, M. L., L. Houzet, et al. (2009). “A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication.“ J Biol Chem 284(29): 19463-73. Huang, X., J. Y. Wang, et al. (2008). “Systems analysis of quantitative shRNA-library screens identifies regulators of cell adhesion.“ BMC Syst Biol 2: 49. Hattori, H., X. Zhang, et al. (2007). “RNAi screen identifies UBE2D3 as a mediator of all-trans retinoic acid-induced cell growth arrest in human acute promyelocytic NB4 cells.“ Blood 110(2): 640-50. Hwang, G. W., T. Hayashi, et al. (2007). “siRNA-mediated inhibition of phosphatidylinositol glycan Class B (PIGB) confers resistance to methylmercury in HEK293 cells.“ J Toxicol Sci 32(5): 581-3. shRNA Library Reviews: Martin, S. E. and N. J. Caplen (2007). “Applications of RNA interference in mammalian systems.“ Annu Rev Genomics Hum Genet 8: 81-108. Echeverri, C. J. and N. Perrimon (2006). “High-throughput RNAi screening in cultured cells: a user's guide.“ Nat Rev Genet 7(5): 373-84. [email protected] 24 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Moffat, J. and D. M. Sabatini (2006). “Building mammalian signalling pathways with RNAi screens.“ Nat Rev Mol Cell Biol 7(3): 177-87. Voorhoeve, P. M. and R. Agami (2003). “Knockdown stands up.“ Trends Biotechnol 21(1): 2-4. Genetic Screens with siRNA libraries: Aza-Blanc, P., Cooper, C.L., Wagner, K., Batalov, S., Deveraux, Q.L. and Cooke, M.P. (2003) Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Molecular Cell. 12: 627-637. Bailey, S.N., Ali, S.M., Carpenter, A.E., Higgins, C. and Sabatini, D. (2006) Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nature Methods. 3: 117-122. Berns, K., Hijmans, E.M., Mullenders, J. et al. (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 428: 431-437. Bortone, K., Michiels, F., Vandeghinste, N., Tomme, P. and van Es, P. (2004) Functional screening of viral siRNA libraries in human primary cells. Drug Discovery World. Fall: 20-27. Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. (2006) An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2(4):202-206. Cullen LM, Arndt GM. (2005) Genome-wide screening for gene function using RNAi in mammalian cells. Immunol Cell Biol. 83:217-23. Devi G. R. siRNA-based approaches in cancer therapy. Cancer Gene Therapy 13: 819-829, 2006. Downward, J. (2004) Use of RNA interference libraries to investigate oncogenic signaling in mammalian cells. Oncogene. 23: 8376-8383. Eggert, U.S., Kiger, A.A., Richter, C., Perlman, Z.E., Perrimon, N., Mitchison, T.J. and Field, C.M. (2004) Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLOS Biology. 2: 1-8. Friedman, A. and Perrimon, N. (2004) Genome-wide high-throughput screens in functional genomics. Cur.Opin.Genet. Develop. 14: 470-476. Huesken, D., Lange, J., Mickanin, C., et al. (2005) Design of a genome-wide siRNA library using an artificial neural network. Nature Biotechnol. 23: 995-1001. Leung RK, Whittaker PA (2005) RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther.107: 222-39. Liang Z. (2005) High-throughput screening using genome-wide siRNA libraries. IDrugs. 11: 924-926. Moffat J, Sabatini DM. (2006) Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol. 7:177-187. Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124(6):1283-98. Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. (2006) A loss-offunction RNA interference screen for molecular targets in cancer. Nature. Mar 29. Paddison, J.P., Silva, J.M., Conklin, D.S. et al. (2004) A resource for large-scale RNA-interference-based screens in mammals. Nature. 428: 427-431. Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 37:1281-8. Poulin G, Nandakumar R, Ahringer J. (2004) Genome-wide RNAi screens in Caenorhabditis elegans: impact on cancer research. Oncogene. 23: 8340-8345. [email protected] 25 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Sachse C, Echeverri CJ.(2004) Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene.23: 8384-8391. Sachse, C., Krausz, E., Kronke, A. et al. (2005) High-throughput RNA interference strategies for target discovery and validation by using synthetic short interfering RNAs: Functional genomic investigations of biological pathways. Methods in Enzymology. 392: 242-277. Silva, J., Chang, K., Hannon, G.J. and Rivas, F.V. (2004) RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene. 23: 8401-8409. Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 37(11):1281-8. Sugimoto A. (2004) High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 72:81-91. Vanhecke, D. and Janitz, M. (2005) Functional genomics using high-throughput RNA interference. Drug. Discov. Today. 10: 205-212. Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. (2006) A genetic screen implicates miRNA372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 124(6):1169-81. Willingham AT, Deveraux QL, Hampton GM, Aza-Blanc P. (2004) RNAi and HTS: exploring cancer by systematic loss-of-function. Oncogene. 23(51):8392-400. Zheng, L., Liu, J., Batalov, S., Zhou, D., Orth, A., Ding, S. and Schultz, P. (2004) Proc. Natl. Acad. Sci. 101: 135140. Viability Screens and Synthetic Lethality References: Reinhardt, H. C., H. Jiang, et al. (2009). “Exploiting synthetic lethal interactions for targeted cancer therapy.“ Cell Cycle 8(19): 3112-9. McLaughlin-Drubin, M. E. and K. Munger (2009). “Oncogenic activities of human papillomaviruses.“ Virus Res 143(2): 195-208. Luo, J., M. J. Emanuele, et al. (2009). “A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene.“ Cell 137(5): 835-48. Scholl, C., S. Frohling, et al. (2009). “Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells.“ Cell 137(5): 821-34. Jiang, H., H. C. Reinhardt, et al. (2009). “The combined status of ATM and p53 link tumor development with therapeutic response.“ Genes Dev 23(16): 1895-909. Boudreau, R. L., I. Martins, et al. (2009). “Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo.“ Mol Ther 17(1): 169-75. Luo, B., H. W. Cheung, et al. (2008). “Highly parallel identification of essential genes in cancer cells.“ Proc Natl Acad Sci U S A 105(51): 20380-5. Kassner, P. D. (2008). “Discovery of novel targets with high throughput RNA interference screening.“ Comb Chem High Throughput Screen 11(3): 175-84. Schwartz, G. F., K. S. Hughes, et al. (2008). “Proceedings of the international consensus conference on breast cancer risk, genetics, & risk management, April, 2007.“ Cancer 113(10): 2627-37. Elliott, D. D., S. I. Sherman, et al. (2008). “Growth factor receptors expression in anaplastic thyroid carcinoma: potential markers for therapeutic stratification.“ Hum Pathol 39(1): 15-20. Fakhoury, J., G. A. Nimmo, et al. (2007). “Harnessing telomerase in cancer therapeutics.“ Anticancer Agents Med Chem 7(4): 475-83. [email protected] 26 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Martin, S. E. and N. J. Caplen (2007). “Applications of RNA interference in mammalian systems.“ Annu Rev Genomics Hum Genet 8: 81-108. Carpten, J. D., A. L. Faber, et al. (2007). “A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.“ Nature 448(7152): 439-44. Echeverri, C. J. and N. Perrimon (2006). “High-throughput RNAi screening in cultured cells: a user's guide.“ Nat Rev Genet 7(5): 373-84. Moffat, J. and D. M. Sabatini (2006). “Building mammalian signalling pathways with RNAi screens.“ Nat Rev Mol Cell Biol 7(3): 177-87. Moffat, J., D. A. Grueneberg, et al. (2006). “A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen.“ Cell 124(6): 1283-98. Brummelkamp, T. R., A. W. Fabius, et al. (2006). “An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors.“ Nat Chem Biol 2(4): 202-6. Shabalina, S. A., A. N. Spiridonov, et al. (2006). “Computational models with thermodynamic and composition features improve siRNA design.“ BMC Bioinformatics 7: 65. Taxman, D. J., L. R. Livingstone, et al. (2006). “Criteria for effective design, construction, and gene knockdown by shRNA vectors.“ BMC Biotechnol 6: 7. Schilling, S. H., M. B. Datto, et al. (2006). “A phosphatase controls the fate of receptor-regulated Smads.“ Cell 125(5): 838-40. Massague, J. and R. R. Gomis (2006). “The logic of TGFbeta signaling.“ FEBS Lett 580(12): 2811-20. An, D. S., F. X. Qin, et al. (2006). “Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors.“ Mol Ther 14(4): 494-504. Birmingham, A., E. M. Anderson, et al. (2006). “3' UTR seed matches, but not overall identity, are associated with RNAi off-targets.“ Nat Methods 3(3): 199-204. Cullen, B. R. (2006). “Induction of stable RNA interference in mammalian cells.“ Gene Ther 13(6): 503-8. Olson, A., N. Sheth, et al. (2006). “RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs.“ Nucleic Acids Res 34(Database issue): D153-7. Farmer, H., N. McCabe, et al. (2005). “Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy.“ Nature 434(7035): 917-21. MacKeigan, J. P., L. O. Murphy, et al. (2005). “Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance.“ Nat Cell Biol 7(6): 591-600. Silva, J. M., M. Z. Li, et al. (2005). “Second-generation shRNA libraries covering the mouse and human genomes.“ Nat Genet 37(11): 1281-8. Huesken, D., J. Lange, et al. (2005). “Design of a genome-wide siRNA library using an artificial neural network.“ Nat Biotechnol 23(8): 995-1001. Boese, Q., D. Leake, et al. (2005). “Mechanistic insights aid computational short interfering RNA design.“ Methods Enzymol 392: 73-96. Jagla, B., N. Aulner, et al. (2005). “Sequence characteristics of functional siRNAs.“ RNA 11(6): 864-72. Wu, G., M. Xing, et al. (2005). “Somatic mutation and gain of copy number of PIK3CA in human breast cancer.“ Breast Cancer Res 7(5): R609-16. Kaelin, W. G., Jr. (2005). “The concept of synthetic lethality in the context of anticancer therapy.“ Nat Rev Cancer 5(9): 689-98. Sachse, C., E. Krausz, et al. (2005). “High-throughput RNA interference strategies for target discovery and validation by using synthetic short interfering RNAs: functional genomics investigations of biological pathways.“ Methods Enzymol 392: 242-77. Silva, J., K. Chang, et al. (2004). “RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age.“ Oncogene 23(51): 8401-9. [email protected] 27 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Zheng, L., J. Liu, et al. (2004). “An approach to genomewide screens of expressed small interfering RNAs in mammalian cells.“ Proc Natl Acad Sci U S A 101(1): 135-40. Mangeot, P. E., F. L. Cosset, et al. (2004). “A universal transgene silencing method based on RNA interference.“ Nucleic Acids Res 32(12): e102. Du, Q., H. Thonberg, et al. (2004). “Validating siRNA using a reporter made from synthetic DNA oligonucleotides.“ Biochem Biophys Res Commun 325(1): 243-9. Willingham, A. T., Q. L. Deveraux, et al. (2004). “RNAi and HTS: exploring cancer by systematic loss-of-function.“ Oncogene 23(51): 8392-400. Downward, J. (2004). “Use of RNA interference libraries to investigate oncogenic signalling in mammalian cells.“ Oncogene 23(51): 8376-83. Yuan, B., R. Latek, et al. (2004). “siRNA Selection Server: an automated siRNA oligonucleotide prediction server.“ Nucleic Acids Res 32(Web Server issue): W130-4. Zhao, J. J., O. V. Gjoerup, et al. (2003). “Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase.“ Cancer Cell 3(5): 483-95. Martin, A. C. and D. G. Drubin (2003). “Impact of genome-wide functional analyses on cell biology research.“ Curr Opin Cell Biol 15(1): 6-13. Voorhoeve, P. M. and R. Agami (2003). “Knockdown stands up.“ Trends Biotechnol 21(1): 2-4. Aza-Blanc, P., C. L. Cooper, et al. (2003). “Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening.“ Mol Cell 12(3): 627-37. Brummelkamp, T. R., S. M. Nijman, et al. (2003). “Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB.“ Nature 424(6950): 797-801. Kumar, R., D. S. Conklin, et al. (2003). “High-throughput selection of effective RNAi probes for gene silencing.“ Genome Res 13(10): 2333-40. Hahn, W. C., S. K. Dessain, et al. (2002). “Enumeration of the simian virus 40 early region elements necessary for human cell transformation.“ Mol Cell Biol 22(7): 2111-23. Tuschl, T. (2002). “Expanding small RNA interference.“ Nat Biotechnol 20(5): 446-8. Hannon, G. J. (2002). “RNA interference.“ Nature 418(6894): 244-51. Vogel, C. L., M. A. Cobleigh, et al. (2002). “Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer.“ J Clin Oncol 20(3): 719-26. Naidu, R., N. A. Wahab, et al. (2002). “Expression and amplification of cyclin D1 in primary breast carcinomas: relationship with histopathological types and clinico-pathological parameters.“ Oncol Rep 9(2): 409-16. Elenbaas, B., L. Spirio, et al. (2001). “Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells.“ Genes Dev 15(1): 50-65. Ross, J. S. and J. A. Fletcher (1998). “The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy.“ Stem Cells 16(6): 413-28. Steck, P. A., M. A. Pershouse, et al. (1997). “Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers.“ Nat Genet 15(4): 356-62. Seshadri, R., C. S. Lee, et al. (1996). “Cyclin DI amplification is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis.“ Clin Cancer Res 2(7): 1177-84. McCann, A. H., A. Kirley, et al. (1995). “Amplification of the MDM2 gene in human breast cancer and its association with MDM2 and p53 protein status.“ Br J Cancer 71(5): 981-5. Marchetti, A., F. Buttitta, et al. (1995). “mdm2 gene alterations and mdm2 protein expression in breast carcinomas.“ J Pathol 175(1): 31-8. [email protected] 28 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net Champeme, M. H., I. Bieche, et al. (1994). “Int-2/FGF3 amplification is a better independent predictor of relapse than c-myc and c-erbB-2/neu amplifications in primary human breast cancer.“ Mod Pathol 7(9): 900-5. Borg, A., B. Baldetorp, et al. (1992). “c-myc amplification is an independent prognostic factor in postmenopausal breast cancer.“ Int J Cancer 51(5): 687-91. [email protected] 29 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net O. Appendix 1. Lentiviral shRNA Expression Vector Maps* (DECIPHER, Standard, and Customizable) [email protected] 30 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net RSV 5’LTR Cellecta pRSI shRNA Expression Lentiviral Vector. AmpR Fully Customizable. RRE pRSI-U6-(sh)-UbiCRFP-2A-Puro ~7.5 - 8.5 kb pUC ORI U6 cPPT (Default features shown) shRNA promoter: BbsI U6 H1 BbsI U6-Tet H1-Tet UbiC SV40 ORI SV40 PolyA Marker promoter: RFP 2A PuroR Markers: for constitutive shRNA expression, any combination of 2 markers (GFP, RFP, PuroR, BleoR, NeoR, Hygro-HK, etc.) hUbiC hEF1 hPGK CMV CMV/Ferritin for tet-regulated shRNA expression, any combination of Tet Repressor (TetR) and one or two markers (GFP, RFP, PuroR, BleoR, NeoR, Hygro-HK, etc.) Standard library expression vector used for library construction. Specific variations shown are available with custom libraries. For sequences and cassette designs for other standard library vectors, please visit the Cellecta website: http://www.cellecta.com/resources/vectors or contact Cellecta at [email protected]. * All Cellecta lentiviral vectors, including the DECIPHER vectors, are covered by a lentiviral expression system license owned by Life Technologies Corporation (LTC). See Terms and Conditions. 2. HT Sequencing Primers HTS6 Cassette (e.g. pRSI16-U6-(sh)-HTS6-UbiC-TagRFP-2A-Puro) Amplicon Size, 2nd round PCR: 251 bp Primer Name F2 R2 Gex1-NF2 Gex2-NR2 GexSeqS FwdU6-1 FwdU6-2 Used for 1st Round 1st Round 2nd Round 2nd Round HT Sequencing Standard sequencing Standard sequencing Sequence (IDT preferred) 5’-TCGGATTCGCACCAGCACGCTA-3’ 5’-AGTAGCGTGAAGAGCAGAGAA-3’ 5’-TCAAGCAGAAGACGGCATACGATCGCACCAGCACGCTACGCA-3’ 5’-AATGATACGGCGACCACCGAGAGCACCGACAACAACGCAGA-3’ 5’-AGAGGTTCAGAGTTCTACAGTCCGAA-3’ (HPLC Purified) 5’-CAAGGCTGTTAGAGAGATAATTGGAA-3’ 5’-CCTAGTACAAAATACGTGACGTAGAA-3’ HTS4 Cassette (e.g. DECIPHER pRSI12-U6-(sh)-HTS4-UbiC-TagRFP-2A-Puro) Amplicon Size, 2nd round PCR: 255 bp Primer Name Used for FwdHTS (was FwdHTS2) RevHTS1 FwdGex (was Gex1MS) RevGex (was Gex2M) GexSeqS FwdU6-1 FwdU6-2 1st Round 1st Round 2nd Round 2nd Round HT Sequencing Standard sequencing Standard sequencing [email protected] Sequence (IDT preferred) 5’-TTCTCTGGCAAGCAAAAGACGGCATA-3’ 5’-TAGCCAACGCATCGCACAAGCCA-3’ 5’-CAAGCAGAAGACGGCATACGAGA-3’ 5’-AATGATACGGCGACCACCGAGA-3’ 5’-AGAGGTTCAGAGTTCTACAGTCCGAA-3’ (HPLC Purified) 5’-CAAGGCTGTTAGAGAGATAATTGGAA-3’ 5’-CCTAGTACAAAATACGTGACGTAGAA-3’ 31 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net HTS3 Cassette (DECIPHER pRSI9-U6-(sh)-HTS3-UbiC-TagRFP-2A-Puro-dW): Amplicon Size, 2nd round PCR: 106 bp Primer Name FwdHTS (was FwdHTS2) RevHTS (was RevcPPT-5) FwdGex (was Gex1MS) RevGex (was Gex2M) GexSeqN FwdU6 (was Fwd-U6-1) FwdU6-2 Used for 1st Round 1st Round 2nd Round 2nd Round HT Sequencing Standard sequencing Standard sequencing Sequence (IDT preferred) 5’-TTCTCTGGCAAGCAAAAGACGGCATA-3’ 5’-TGCCATTTGTCTCGAGGTCGAGAA-3’ 5’-CAAGCAGAAGACGGCATACGAGA-3’ 5’-AATGATACGGCGACCACCGAGA-3’ 5’-ACAGTCCGAAACCCCAAACGCACGAA-3’ (HPLC Purified) 5’-CAAGGCTGTTAGAGAGATAATTGGAA-3’ 5’-CCTAGTACAAAATACGTGACGTAGAA-3’ To verify the HT sequencing primers for other standard library vectors, please visit the Cellecta website: http://www.cellecta.com/resources/vectors/ or contact Cellecta at [email protected]. 3. Common Library Vector Features Feature Rous Sarcoma Virus (RSV) enhancer/promoter HIV-1 truncated 5′ LTR HIV-1 psi (ψ) packaging signal HIV-1 Rev response element (RRE) U6 cPPT UbiC promoter TagRFP 2A (T2A) PuroR WPRE ∆U3/HIV-1 truncated 3′ LTR SV40 polyadenylation signal SV40 Ori AmpR pUC ori Function Source Allows Tat-independent production of viral mRNA (Dull et al., 1998). Rous sarcoma virus Permits viral packaging and reverse transcription of the viral mRNA (Luciw, 1996). HIV-1 Allows viral packaging (Luciw, 1996). HIV-1 Permits Rev-dependent nuclear export of unspliced viral mRNA (Kjems et al., 1991; Malim et al., 1989). Human U6 promoter drives RNA Polymerase III transcription for generation of shRNA transcripts. Central polypurine tract, cPPT, improves transduction efficiency by facilitating nuclear import of the vector's preintegration complex in the transduced cells. Ubiquitin C promoter drives expression of TagRFP and PuroR. TagRFP fluorescent protein (Evrogen) serves as an indicator of successful transduction. Thosea asigna virus 2A translational cleavage site containing 18 amino acid residues. Cleavage occurs via a cotranslational ribosome skipping mechanism between the Cterminal glycine and proline residues, leaving 17 residues attached to the end of TagRFP and 1 residue to the start of the puromycin resistance marker (in the DECIPHER vectors). Puromycin-resistant marker for selection of the transduced cells. Woodchuck hepatitis virus posttranscriptional regulatory element—enhances the stability of viral transcripts. 3' Self-inactivating long terminal repeat. Allows viral packaging but self-inactivates the 5′ LTR for biosafety purposes (Dull et al., 1998). The element also contains a polyadenylation signal for transcription termination and polyadenylation of mRNA in transduced cells. Required for viral reverse transcription; self- inactivating 3' LTR with deletion in U3 region prevents formation of replicationcompetent viral particles after integration into genomic DNA. Allows transcription termination and polyadenylation of mRNA. Allows for episomal replication of plasmid in eukaryotic cells. Ampicillin resistance gene (β-lactamase) for selection of plasmid in bacterial cells. pUC bacterial origin of replication. HIV-1 Human HIV-1 Human sea anemone Entacmaea quadricolor Thosea asigna virus Streptomyces alboniger Woodchuck hepatitis virus HIV-1 SV40 SV40 bacterium Salmonella paratyphi pUC * (c): element on complementary strand [email protected] 32 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net 4. DECIPHER Library HT Sequencing Q.C. Data Complete Plasmid shRNA Library HT sequencing data for all modules is available on the DECIPHER Project website at http://www.decipherproject.net/support/#sequences. Plasmid HT Sequencing data may be used as negative control (untreated/untransduced/day 0) data for many types of genetic screens. The shRNA/barcode representation histograms for individual DECIPHER libraries are available on the PAC forms available on the Cellecta website at http://www.cellecta.com/resources/protocols/. 5. DECIPHER Library Individual Clone Sequencing Q.C. Data DECIPHER Libraries in pRSI12 Vector: Human M1 Human M2 Human M3 Mouse M1 Mouse M2 Lot #: DECIPHER Library: 11070805 12052001 12052002 13011802 13011803 Library Complexity (number of clones): >50 × 10 90 × 10 180 × 10 n/a n/a Number of random clones picked: Single Insert Rate: Number of clones with at least one mutation, deletion, or insertion: Mutation / Deletion / Insertion Rate: Estimated % of Inserts without any mutations, deletions, or insertions in antisense portion and considered to be functional: 6 6 6 40 24 24 24 19 >95% >95% >95% >95% >95% 2 3 5 2 1 0.1 – 0.2% 0.25% 0.32% 0.15% 0.1% >95% 95% 93% 95% 95% 6. DECIPHER Project Resources DECIPHER Library users have access to additional valuable resources. Barcode Analyzer and Deconvoluter http://www.decipherproject.net/software/#barcode-deconvoluter This software is required to convert raw HT sequencing data from DECIPHER library screens into a summary file for subsequent processing, and it includes annotation for every identified gene. Next, data can be processed, edited, normalized, and transformed using your data analysis tool of choice, such as SAS, SPSS, or, for simpler analyses, Microsoft Excel. The program requires the appropriate DECIPHER Module Library (BLIB) files, which are available for download at http://www.decipherproject.net/software/#blib-files. To install, just move them to the same directory where the Barcode Deconvoluter software resides. View the Frequently Asked Questions (FAQ) page here: http://www.decipherproject.net/support/frequently-asked-questions/. List of Functionally Validated shRNA http://www.decipherproject.net/software/#validated-shrna-sequences As a result of NIH SBIR grants HG003355 and RR024323, Cellecta has compiled a database of ~120,000 functionally validated shRNA for human and mouse genes. Under the NIH Data and Resource Sharing Plan, the shRNA sequences are freely available to all academic and commercial researchers. RNAi Generator Tool http://www.decipherproject.net/software/#third-party Generate a list of optimal shRNA target sequences for given input sequences. The software is extremely customizable. Authored by Gus Frangou, Ph.D. of the Roswell Park Cancer Institute. [email protected] 33 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net P. Terms and Conditions Cellecta, Inc. Limited License Cellecta grants the end user (the “Recipient”) of the Pooled Lentiviral shRNA Libraries and Vector (the “Product”) a nontransferable, non-exclusive license to use the reagents for internal research use only as described in the enclosed protocols; in particular, research use only excludes and without limitation, resale, repackaging, or use for the making or selling of any commercial product or service without the written approval of Cellecta, Inc. -- separate licenses are available for non-research use or applications. The Product is not to be used for human diagnostics or included/used in any drug intended for human use. Care and attention should be exercised in handling the Product by following appropriate research laboratory practices. Cellecta’s liability is expressly limited to replacement of Product or a refund limited to the actual purchase price. Cellecta’s liability does not extend to any damages arising from use or improper use of the Product, or losses associated with the use of additional materials or reagents. This limited warranty is the sole and exclusive warranty. Cellecta does not provide any other warranties of any kind, expressed or implied, including the merchantability or fitness of the Product for a particular purpose. Use of the Product for any use other than described expressly herein may be covered by patents or subject to rights other than those mentioned. Cellecta disclaims any and all responsibility for injury or damage that may be caused by the failure of the Recipient or any other person to use the Product in accordance with the terms and conditions outlined herein. The Recipient may refuse these licenses by returning the enclosed Product unused. By keeping or using the enclosed Product, you agree to be bound by the terms of these licenses. The laws of the State of California shall govern the interpretation and enforcement of the terms of these Licenses. DECIPHER™ shRNA Libraries Cellecta’s Plasmid DECIPHER shRNA Libraries are covered under a fully executed Materials Transfer Agreement (MTA) between Cellecta and the receiving institution. Please visit http://www.decipherproject.net/support/#ordering or contact the DECIPHER Project Manager at [email protected] for more information. If purchased from Cellecta at list price, Plasmid DECIPHER shRNA Libraries do not require nor are subject to the conditions of an MTA. Limited Use Label Licenses The Recipient acknowledges that the Product has been developed by Cellecta based on licenses from Third Parties and agrees with the Terms of Limited Use for the Recipient provided by the Third Parties: Agilent Technologies, Inc. End-User Label License for the use of shRNA libraries comprising Oligo Pools: “This Internal Use only license grants End-Users the sole right to use and fully consume or destroy this product (the “Product”). Use of the Product is limited to Research Use ONLY, not for diagnostic procedures. In all cases, sale or other transfer or distribution to third parties of (i) the Product, or any portion, (ii) DNA, RNA and protein constructs or libraries created from the Product or any portion, or of (iii) transformed phage, viruses, cells, or tissues created directly or indirectly from the Product, or any portion is strictly prohibited without prior written approval by Agilent Technologies, Inc.” Life Technologies Corporation End-User Label License for the use of Lentiviral Expression System: “This product or service (based upon the Lentiviral Expression System) is sublicensed from Life Technologies Corporation under U.S. Patent Nos. 5,686,279; 5,834,256; 5,858,740; 5,994,136; 6,013,516; 6,051,427; 6,165,782; 6,218,187; 6,428,953; 6,924,144; 7,083,981 and 7,250,299 and corresponding patents and applications in other countries for internal research purposes only. Use of this technology for gene therapy applications or bioprocessing other than for nonhuman research use requires a license from GBP IP, LLC. Please contact GBP IP, LLC 537 Steamboat Road, Suite 200, Greenwich, CT 06830. Use of this technology to make or sell products or offer services for consideration in the research market requires a license from Life Technologies Corporation, 5791 Van Allen Way, Carlsbad, CA 92008.” Evrogen IP JSC End-User Label License for the use of lentiviral shRNA constructs comprising TagRFP-encoded gene: “This product is for internal non-commercial research use only. No rights are conveyed to modify or clone the gene encoding fluorescent protein contained in this product. The right to use this product specifically excludes the right to validate or screen compounds. For information on commercial licensing, contact Evrogen Licensing Department, email: [email protected]”. Cold Spring Harbor Laboratory (CSHL) End-User Label License for use of expression vectors encoding an shRNA: Acceptance. This Limited Use License (“License”) contains the exclusive terms and conditions between CSHL and Customer for use of the Product. By opening the Product container or in any other way accessing or using the Product (“Acceptance”), you will create a binding legal contract upon the terms and conditions herein, without modification. Customer's purchase order or similar terms shall not apply to this License. If you are not authorized by Customer to enter into this License or do not agree to all terms and conditions in this License, then you are prohibited from opening the Product container or otherwise accessing or using the Product. Permitted Use. Portions of the Product are covered by US and foreign patent applications or patents and other proprietary intellectual property rights owned by CSHL (“shRNA IP Rights”). Subject to Acceptance and all terms and conditions of this License, sale of the Product to Customer by Seller (acting under its license from CSHL, an “Authorized Sale”) conveys to Customer only the nonexclusive, nontransferable right under the shRNA IP Rights to use the Product solely for Customer's internal research purposes, and only at its facility where the Products are delivered by Seller. Unlicensed Products. Any Product that is acquired other than pursuant to an Authorized Sale (including without limitation, any Product not acquired from Seller) shall be deemed to be an “Unlicensed Product”. This License shall be void and of no effect for Unlicensed Products and shall not convey any express or implied right to make, use or sell Unlicensed Products for any purpose. Restrictions. Customer obtains no right to sublicense it rights, or to use the Product for the benefit of any third party for any commercial purpose (including without limitation, using the Product in connection with providing services to any third party or [email protected] 34 of 35 v.5f, 7/3/14 Cellecta Pooled Lentiviral shRNA Libraries User Manual www.cellecta.com www.decipherproject.net generating commercial databases). The Product may not be used in vitro or in vivo for any diagnostic, preventative, therapeutic or vaccine application, or used (directly or indirectly) in humans for any purpose. Customer may not isolate, extract, reverse engineer, derive, copy or separately use any component of the Product (such as, for example, any shRNA component) for any commercial purpose (including without limitation, for the purpose of making Products) other than solely for Customer's internal research purposes. Non-Profit Customers. If Customer is a Non-Profit Entity, then the following additional restrictions shall apply: Customer obtains no right to use the Product for any commercial purpose. Commercial Customers. If Customer is a Commercial Entity, (unless Customer has already entered into a separate written agreement that has been executed by CSHL, that covers the shRNA IP rights, and that is then currently in effect) then the following additional restrictions shall apply: This License and Customer’s rights hereunder automatically terminate 1 year after delivery of Product to Customer. After 1 year of Product use customer must enter into a separate written agreement with CSHL that covers the shRNA IP rights or Customer shall immediately stop using and destroy all Product in its possession. The Product may not be used to make any mouse that is of a strain of mice for germ line transmission by embryonic transfer of a gene encoding an shRNA that induces suppression of a gene or genes by RNAi. No Transfers. Customer may not distribute or transfer the Product (by license, sale, loan, lease, rental or any other means) to any commercial partner or any other third party for any commercial purpose, except only in the following case. Customer may transfer the unmodified Product to a commercial third party contractor who, pursuant to a written agreement with Customer and only for non-royalty based payment(s), undertakes on behalf of Customer to use the Product solely for Customer’s benefit and internal research purposes, which third party shall not, after termination of such work, retain or receive subsequent rights to possess, access or use any Product (or any results of such work), and from whom Customer receives no payments pursuant to such agreement. Compliance. Customer may only use the Product in compliance with all local, state, federal and other applicable laws, regulations and rules, including without limitation (for uses in the United States), EPA, FDA, USDA and NIH guidelines, Customer may not (directly or indirectly) use the Product, or allow the transfer, transmission, export or re-export of all or any part of the Product or any product thereof, in violation of any export control law or regulation of the United Sates or any other relevant jurisdiction. Disclaimers. THE PRODUCT IS PROVIDED “AS IS” WITHOUT WARRANTY OF ANY KIND. NO WARRANTY IS MADE THAT THE PRODUCT WILL MEET CUSTOMER’S REQUIREMENTS, OR THAT ANY RESULT CAN BE ACHIEVED, OR THAT USE OF THE PRODUCT WILL NOT INFRINGE ANY PATENT OR OTHER PROPRIETARY RIGHT. ALL WARRANTIES, EXPRESS OR IMPLIED, ORAL OR WRITTEN, ARE HEREBY EXPRESSLY DISCLAIMED, INCLUDING WITHOUT LIMITATION, ALL IMPLIED WARRANTIES OF NON-INFRINGEMENT, QUIET ENJOYMENT, MERCHANTABILITY OR FITNESS FOR ANY PARTICULAR PURPOSE AND ALL WARRANTIES ARISING FROM ANY COURSE OF DEALING, COURSE OF PERFORMANCE OR USAGE OF TRADE. Other Uses. Except for the limited use expressly specified above, no other license is granted, no other use is permitted and CSHL retains all rights, title and interests in and to the shRNA IP Rights. Nothing herein confers to Customer (by implication, estoppel or otherwise) any right or license under any patent, patent application or other proprietary intellectual property right of CSHL other than the shRNA IP Rights. For information on purchasing a license to use the Product for longer time periods, in greater quantities or for other purposes, or to practice more broadly under the shRNA IP Rights, or to practice under other CSHL intellectual property rights, please contact the CSHL Office of Technology Transfer at (516) 367-8301. Definitions. “Affiliate” means, at the time of reference thereto, any corporation, company, partnership, joint venture or other entity which controls, is controlled by or is under common control with the subject entity, where “control” means direct or indirect ownership of more than 50% of (i) the outstanding stock or other voting rights entitled to elect directors or (ii) all ownership interests (or, in any country where the local law shall not permit foreign equity participation of 50% or more, then the direct or indirect ownership or control of the maximum percentage of such outstanding stock, voting rights or ownership interests permitted by local law). “Commercial Entity” means any entity or organization other than a Non-Profit Entity. “CSHL” means Cold Spring Harbor Laboratory. “Customer” means the company or other entity or organization that orders, pays for and takes delivery of the Product. “Non-Profit Entity” means any college, university or governmental entity (including without limitation, governmental and quasi-governmental institutes and research laboratories), or any non-profit scientific, research or educational organization that is of the type described in section 501(c)(3) of the Internal Revenue Code or that is qualified under a state non-profit organization statute. “Product” means a product (including without limitation, expression vectors encoding an shRNA), the design, manufacture or use of which (in whole or in part) is the subject of the shRNA IP Rights, and is deemed to include all components, progeny, reproductions, modified versions and other derivatives thereof. “Seller” means Cellecta. Inc. © 2014 Cellecta, Inc. All Rights Reserved. Trademarks CELLECTA is a registered trademark of Cellecta, Inc. DECIPHER is a trademark of Cellecta, Inc. CRL-11268 is a trademark of ATCC. Invitrogen, Lipofectamine, and Plus Reagent are trademarks of Life Technologies Corporation. [email protected] 35 of 35 v.5f, 7/3/14
© Copyright 2026