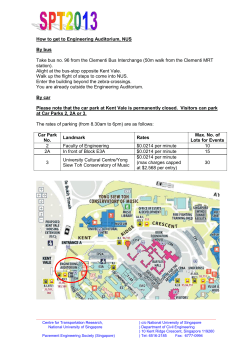
MERLION Metabolomics Workshop Singapore 2014 Workshop Programme
MERLION Metabolomics Workshop Singapore 2014 19 – 21 November 2014 University Town, NUS, Singapore Developing Metabolomics Platform Technologies through Singapore-French Research Alliance Workshop Programme And Abstract Book Organised by: MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 CONTENT Content Page ...............................................................................................1 Welcome Message ......................................................................................2 Organisers’ Welcome ..................................................................................3 Organising Committee ................................................................................4 NUS Environmental Research Institute .................................................... 5 MetaboHUB .................................................................................................6 Institut Français de Singapour – French Embassy ................................... 7 Keynote Speakers........................................................................................8 Programme ................................................................................................13 Young Researcher Award .........................................................................21 Abstracts for Main Session.......................................................................22 Abstracts for Young Researchers Session ...............................................55 Agilent Sponsored Talk .............................................................................73 Technical Site Visit ....................................................................................74 Sponsors’ profiles ......................................................................................76 Location Map of University Town ...........................................................79 Notes ..........................................................................................................81 1 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Welcome Message from the Organising Committee Chairperson As the Chairperson, it is my pleasure to welcome you to the Merlion Metabolomics Workshop Singapore 2014. This is the first 3-day workshop jointly organized by Singapore and France, providing first-hand insights and experiences revolving around 3 broad themes of metabolomics – Human Health, Food and Science Technology and Environmental. Through this workshop, we hope to encourage technical exchanges and catalyze the development of research collaboration in the field of metabolomics between Singapore and France. Apart from Singapore and France, we have participants from Australia, China, Japan, Hong Kong, South Korea, India, Ireland, Thailand, Malaysia, Czech Republic, Italy and United Kingdom. This workshop is truly an epitome of Singapore’s identity, a cosmopolitan melting pot, where east meets west. We hope to encourage the transfer of knowledge and technologies developed in Europe to Asia and vice versa. To end, I would like to express my personal gratitude to faculty and staff at the National University of Singapore, Metabohub and the Institut Français de Singapour – French Embassy, industry partners and sponsors for their work and dedication. Also to participants, thank you for helping us make this Merlion Metabolomics Workshop Singapore 2014 a success. Sincerely, Chairperson, MMWS 2014 Associate Professor, Department of Biological Sciences, NUS Deputy Director, NUS-NERI Director, Graduate Program, NTU-SCELSE 2 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Organisers’ Welcome Dear Participants, We would like to extend a warm welcome to everyone attending the 2014 Merlion Metabolomics Workshop Singapore! We have received an overwhelming response from the scientific community. We would like to thank every invited speakers and participants for attending this workshop. Your enthusiastic participation ensures the very success of this workshop. This Merlion Metabolomics workshop has an exciting three days programme addressing areas of Human Health, Food and Science Technology and Environmental Metabolomics. The goal of this workshop is to establish an open channel of communication, networking and to encourage mutual sharing of ongoing research results in the field of metabolomics between Singapore and France. At the end of each day, we have sessions for young researchers to share their research findings. We hope that this will be an enjoyable and interactive event for all. Enjoy your stay in Singapore! Best wishes to all delegates, Organising Committee MMWS2014 3 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Organising Committee Organising Committee Associate Professor Sanjay SWARUP, National University of Singapore Professor Choon Nam ONG, National University of Singapore Professor Sam Fong Yau LI, National University of Singapore Associate Professor Eric Chun Yong CHAN, National University of Singapore Dr. Fabien JOURDAN, French National Research Institute for Agricultural Research Ms. Caroline SAUTOT, French National Research Institute for Agricultural Research Transfert Mr. Geoffry SMITH, International Life Sciences Institute Southeast Asia Mr. Florent BEAU, Institut Français de Singapour – French Embassy Dr. Adeline MARTIN, Institut Français de Singapour – French Embassy Dr. Peter KEW, National University of Singapore Ms. Elaine TAY, National University of Singapore Ms. Eileen Mei Hui TAN, National University of Singapore Scientific Committee Associate Professor Sanjay SWARUP, National University of Singapore Professor Dominique ROLIN, University of Bordeaux Dr. Fabien JOURDAN, French National Research Institute for Agricultural Research Professor Sam Fong Yau LI, National University of Singapore Mr. Geoffry SMITH, International Life Sciences Institute Southeast Asia 4 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 About NUS Environmental Research Institute (NERI) The NUS Environmental Research Institute (NERI) aims to be a leading global institute for interdisciplinary research, education and expertise in environmental science and technology in Asia. NERI coordinates and facilitates cross-faculty research and educational activities to conduct cutting-edge research addressing critical environmental issues. NERI proactively engages strategic partners within NUS' global networks, with government agencies, and industry. The current key research tracks are: • • • • Environmental Surveillance and Treatment Environmental and Human Health Green Chemistry and Sustainable Energy Impacts of Climate Change on the Environment For more information, visit http://www.nus.edu.sg/neri/. FOR A SUSTAINABLE FUTURE 5 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 About Metabohub MetaboHUB is the French infrastructure of metabolomics and fluxomics that provides tools and services to academic research teams and industrial partners in the fields of health, nutrition, agriculture, environment and biotechnology. MetaboHUB infrastructure gathers the equipment, means, competences, and critical mass to respond to the specific requirements of current and future metabolomics: multi-tools requirement, large-scale analysis programs, standardization of data and databases establishment, multicompetences requirements and systems biology approach. MetaboHUB provides a large variety of metabolomics services and expertise to your researcher. The main services consist in analyzing of many biological matrices using the tools of metabolomics and fluxomics. Our laboratories have the expertise and knowledge of sample preparation and data acquisition from a wide range of biological matrices (plants, microorganisms, human cells, animal tissues and biofluids). These services include data acquisition by mass spectrometry (GC, LC), NMR (liquid and HR-MAS) and LC-NMR, NMR/MS hyphenation and processing, annotation, visualization and exploration of large data set. A team of bioinformaticians and biostatisticians is supporting the achievement of these remaining tasks. The advanced services cores offers collaborations on a project-basis in specific areas of metabolism from targeted metabolites analysis including primary, secondary metabolites, lipids, xenobiotics, drugs, etc. to non-targeted metabolomics including lipidomics. MetaboHUB hosts a comprehensive park of analytical tools and owns a strong expertise for the quantification of a wide range of metabolites. MetaboHUB is committed to transfer its technical knowledge and communicate on breakthroughs to the broad scientific community (from classroom to lab bench). MetaboHUB partners already propose a wide range of personalized training sessions to the scientific community upon request (e.g. in metabolomics and fluxomics; in data treatment and biostatistics; in NMR profiling, database management, data curation). MetaboHUB staff members are already committed to knowledge dissemination through the “Réseau Français de Métabolomique et Fluxomique” (‘RFMF’, French Metabolomics and Fluxomics Network) and through "Réseau Français des Lipidomistes" (RFL). Various events are being organized by MetaboHUB staff through RFMF and MetaboHUB is largely involved in the organization of several national and international congresses. For more information, visit: http://www.metabohub.fr/en/ Contact: [email protected] ANR-11-INBS-0010 6 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 About Institut Français de Singapour – French Embassy The Institut Français de Singapour (IFS) of the French Embassy is a vector that develops cooperation between France and Singapore in the areas of culture, science and technology, and higher education. It promotes the French language, educational opportunities, creativity and French innovation and research. It thus has a far reaching mandate and is a center of expertise and advice. Always at the forefront of current issues, the Institut Français de Singapour’s priority is a focus on innovation, making it a factor of attractiveness for French expertise. Among the numerous programs of the IFS, the PHC Merlion program actively supports research development. The PHC Merlion Program is a joint Franco-Singaporean collaboration, and managed by the IFS in partnership with Singaporean institutions. The aim is to encourage and support new scientific development in research between French and Singaporean laboratories, through funding the scientists’ trip exchanges. The program takes the form of an annual call for proposals, and since its inception in 2006, has funded more than 150 bilateral projects, one of which is the MERLION Metabolomics Workshop Singapore 2014! More information and the calendar of cultural and scientific events may be found on our website. In addition, do not hesitate to subscribe to our newsletter. http://www.institutfrancais.sg/ 7 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Keynote Speakers Professor Choon Nam ONG School of Public Health, National University of Singapore (NUS) Director of the NUS Environmental Research Institute, Singapore His current research interests include evaluation of Biomarkers using metabolomics platform and Mechanistic-based investigations of chronic disease and their prevention. He has published over 300 peerreviewed articles with close to 15,000 citations. He sits on the editorial board of several scientific journals and currently an Associate Editor of Environmental Research. Professor Dominique ROLIN Metabohub – Bordeaux University of Bordeaux, France D. Rolin is a professor at University of Bordeaux. He has recognized expertise in the fields of NMR spectroscopy, biology, plant and fruit metabolism, metabolomics and fluxomics (M&F). He is heading MetaboHUB and also CGFB a federation of technological platforms dedicated to the study of living organisms at University of Bordeaux. In 2002, he has set up and led the Metabolome Facility of Bordeaux (BMP), which can be considered as a pioneer in plant metabolomics in France. D. Rolin is also deeply involved in the promotion of M&F in France as co-founder (2005), treasurer (2007-2010) and President (2010-2015) of the RFMF. 8 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Professor Royston GOODACRE University of Manchester, United Kingdom Educated in University of Bristol, UK where he was awarded his BSc in Microbiology and his PhD in analytical methods applied to microbiological problems. After which he did a three year PDRA in University of Wales, Aberystwyth, UK and then four years as a Wellcome Trust Fellow where he investigated chemometrics and artificial neural networks for the analysis of spectroscopic data. Following this he was appointed a Lecturer in Microbiology. He moved in Feb 2003 to take up the position of Reader in Analytical Science in the School of Chemistry, The University of Manchester, UK. His current position since Aug 2005 is as Professor of Biological Chemistry. His research focuses on developing MS-based metabolomics along with new chemometric approaches that he applies to help understand how organisms respond to external perturbations. His group also develops Raman spectroscopy for chemical imaging and surface enhanced Raman scattering for trace detection of (bio)chemicals. He is founder and Editor-in-Chief of Metabolomics, on the Editorial Advisory Boards of Analyst and Journal of Analytical and Applied Pyrolysis. Associate Professor Robert TRENGOVE Separation Science and Metabolomics Laboratory Murdoch University, Australia Rob Trengove has pioneered the development of MS-based metabolomics techniques for more than 20 years, collaborating with Australian and International researchers and industry. He currently leads a team of more than 15 researchers working on a diverse range of topics including HIV/AIDs, iron disorders, desalination, microalgae lipidomics, fungal and bacterial metabolomics. Rob Trengove has published more than 60 high-impact journal articles and has ongoing commercial contract research arrangements with major industry players in the petrochemical, clinical and animal health pharmaceutical sector. 9 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Professor Huiru Tang Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, China Prof. TANG received his bachelor’s degree in 1986 from Shannxi University of Science and Technology and doctor’s degree in 1994 from University of London. His research interests include metabolomic analytical methods, interaction between host metabolic and intestinal flora, pathophysiological metabonomic studies. He has published more than 130 papers with more than 2000 citations. Professor Jean Charles Portais Metabolic Biochemistry at the University of Toulouse, France JC PORTAIS is Professor of Metabolic Biochemistry at the University of Toulouse (France) and has recognized expertise in the fields of metabolism, metabolomics and fluxomics. He is heading the research group MetaSys at LISBP, which aims at the comprehensive, system-level understanding of metabolic adaptation and its role in the behavior of organisms, with applications in biotechnology and human health. Prof. JC Portais is also heading MetaToul, the Metabolomics & Fluxomics Center of Toulouse, one of the very first Metabolomics centers in France. He is also deeply involved in the promotion of M&F in France as cofounder and President (2005-2010) of the RFMF. 10 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Professor Eiichiro Fukusaki Division of Science and Biotechnology, Graduate School of Engineering, Osaka University, Japan Eiichiro Fukusaki entered a private company, Nitto Denko Co, after receiving master degree from Osaka University on 1985. He received PhD from Osaka University on 1993 through his company work. After ten years company experience, he returned back to Osaka University as an associate professor. On 2007 he has been assigned as a full professor in Osaka University. His current research interests are focusing on development and application of metabolomics technology. He has published over 200 original papers and 50 patents. Dr Christophe Junot French Alternative Energies and Atomic Energy Commission, France C. Junot is a doctor of Pharmacy. After having gained a PhD in Analytical Chemistry in 2000 (Pierre et Marie Curie University, Paris 6), he joined GlaxoSmithKline laboratories and developed experience in the field of pharmacokinetics and metabolism applied to drug discovery for 2 years. From 2002, he has been a group leader at the Life Science Division of CEA (Commissariat à l’Energie Atomique) where he develops mass spectrometry based analytical methodologies for metabolome analysis. C. Junot has been appointed as head of the Laboratory for Drug Metabolism Studies since September 2010 and he is member of the board of the French Metabolomics and Fluxomics Network. C. Junot is the deputy coordinator of the French MetaboHUB platform and also in charge of the coordination of analytical chemistry developments within this platform. 11 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Dr Fabien Jourdan MetaboHub-Toulouse French National Research Institute for Agricultural Research, France Fabien Jourdan is a research scientist at INRA (the French National Research Institute for Agricultural Research). He is developing bioinformatics methods to study genome-scale metabolic networks. These approaches are mainly based on graph models. The aim is, based on experimental data like metabolomics ones, to retrieve parts of the organism metabolism affected by genetic or environmental perturbations. Fabien Jourdan is leading the development of MetExplore web server (www.metexplore.fr) which is gathering most of these algorithms and allows visualization of metabolic networks. MetExplore development is supported by the French National infrastructure for metabolomics and fluxomics, MetaboHub.. 12 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Scientific Programme Date (Day) Time Event 19 Nov 2014 (Wednesday) A1 A2 Speakers Day 1 8.30 - 9.00 a.m. Registration and Breakfast 9.00 - 9.10 a.m. Opening address by National University of Singapore Assoc Prof Sanjay Swarup Chairman Merlion Metabolomics Workshop 9.10 - 9.20 a.m. Welcome address by Invited Guest National University of Singapore Prof Andrew Wee Vice President (University and Global Relations) National University of Singapore 9.20 - 9.30 a.m. Speech by Guest-of-Honor Embassy of France to Singapore His Excellency Mr Benjamin Dubertret Ambassador of France to Singapore 9.30 - 10.00 a.m. Opening Speaker (Singapore) Introducing metabolomics in Singapore Prof Ong Choon Nam NUS Environmental Research Institute National University of Singapore 10.00 - 10.30 a.m. Opening Speaker (France) MetaboHUB and RFMF: Two tools at the service of metabolomics and fluxomics in France Prof Dominique Rolin Metabohub - Bordeaux University of Bordeaux, France 10.30 - 10.50 a.m. Tea break 13 MERLION METABOLOMICS WORKSHOP SINGAPORE 19 Nov 2014 (Wednesday) Abstract No. 2014 Day 1 Session 1 - Food Science and Technology Metabolomics Chaired by: Dr Fabien Jourdan and Dr Naweed Naqvi 10.50 – 11.20 a.m. Keynote Speaker Metabolomics the way forward Prof Royston Goodacre Manchester Institute of Biotechnology University of Manchester, United Kingdom A4 11.20 – 11.50 a.m. Keynote Speaker Metabolic network modelling and Food Toxicology Dr Fabien Jourdan MetaboHub-Toulouse French National Research Institute for Agricultural Research, France A5 11.50 – 12.10 p.m. Untargeted metabolomic approaches and data mining tools for marker discovery in nutrition Dr Estelle Pujos-Guillot French National Research Institute for Agricultural Research, France A6 12.10 – 12.30 p.m. Lipidomic analysis of palm oil variability Dr Laetitia Fouillen French National Center for Scientific Research, France 12.30 - 12.50 p.m. Novel Metabolites Involved in Pathogen-Host Interaction and Disease Control Dr Naweed Naqvi Temasek Life Sciences Laboratory National University of Singapore A3 A7 12.50 - 1.30 p.m. Lunch 14 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 19 Nov 2014 (Wednesday) Day 1 Abstract No. Session 2 – Human Health Metabolomics Chaired by: Dr Christophe Junot and Assoc Prof Chan Chun Yong, Eric 1.30 – 2.00 p.m. Keynote Speaker Mass spectrometry based metabolomics and lipidomics for the study rare diseases Dr Christophe Junot French Alternative Energies and Atomic Energy Commission, France 2.00 – 2.20 p.m. Lipidomics: a key tool for human health Dr Justine Bertrand Michel French National Institute of Health and Medical Research, France 2.20 – 2.40 p.m. Urinary metabolomics revealed arsenic exposurerelated metabolic alteration: a proof-of-concept study in a Chinese male cohort Prof Zhang Jie Institute of Urban Environment Chinese Academy of Sciences, China A11 2.40 – 3.00 p.m. Noninvasive urinary metabonomic diagnosis of human bladder cancer A12 3.00 – 3.20 p.m. Natural variation of lipidomes A8 A9 A10 3.20 - 3.40 p.m. Assoc Prof Chan Chun Yong, Eric Department of Pharmacy National University of Singapore, Singapore Assoc Prof Marcus Wenk Department of Biochemistry and Department of Biological Sciences National University of Singapore, Singapore Tea break Session 3 - Young Researchers Session Y1-Y8 3.40 - 5.40 p.m 5.40 – 6.00 p.m. 6.00 – 8.00 p.m. 8 oral presentations (15 min each) - Human Health Presenters: 1. Dr Mrinal Kumar Das 2. Dr Cui Liang 3. Dr Liu Feng 4. Ms Yin Xuejiao 5. Mr Erhan Simsek 6. Ms Anna Karen Carrasco Laserna 7. Dr Kenjiro Kami 8. Dr Robert L Davidson Chaired By: Dr Sastia Prama Putri Department of Biotechnology Graduate School of Engineering Osaka University, Japan Agilent Sponsored Talk Dr Oliver Jones Two-dimensional liquid School of Applied Sciences, chromatography: a new RMIT University, Australia technique for metabolomics? Welcome Cocktail Reception 15 MERLION METABOLOMICS WORKSHOP SINGAPORE 20 Nov 2014 (Thursday) 2014 Day 2 Session 4 – Food Science and Technology Metabolomics Chaired by: Prof Royston Goodacre and Dr Blandine Comte 9.00 – 9.30 a.m. Keynote Speaker Application of metabolic fingerprinting for food quality assessments Prof Eiichiro Fukusaki Department of Biotechnology Graduate School of Engineering Osaka University, Japan 9.30 – 9.50 a.m. Metabolomics and systems biology in the context of food toxicology Dr Daniel Zalko French National Research Institute for Agricultural Research, France 9.50 – 10.10 a.m. Use of metabolomics for gene function discovery in a dwarf rice mutant A16 10.10 – 10.30 a.m. Nutritional metabolomics: an integrative understanding of metabolic disease development Dr Blandine Comte French National Research Institute for Agricultural Research, France A17 10.30 - 10.50 a.m. A totally new way to measure the germination of grain using metabolomics Dr Oliver Jones School of Applied Sciences, RMIT University, Australia A13 A14 A15 10.50 - 11.10 a.m. Prof Prakash Kumar Department of Biological Sciences National University of Singapore, Singapore Tea break 16 MERLION METABOLOMICS WORKSHOP SINGAPORE 20 Nov 2014 (Thursday) Day 2 Abstract No. Session 5 – Human Health Metabolomics Chaired by: Prof Ong Choon Nam and Dr Etienne Thevenot 2014 A18 11.10 - 11.40 a.m. Keynote Speaker Metabolomic Aspects of Symbiosis and Transgenomic Interactions In Mammals A19 11.40 – 12.00 p.m. Metabolomic applications in Dr Ho Ying Swan Bioprocessing Technology bioprocessing and clinical Institute, A*STAR, Singapore research 12.00 a.m. – 12.20 p.m. Metabolomics driven the discovery of functional small molecules towards a diversity of biological innovations Prof Lu Haitao Chongqing University Innovative Drug and Research Centre, China 12.20 – 12.40 p.m. Urine, metabolite profiling, its applications Dr Ong Eng Shi Singapore University of Technology and Design, Singapore 12.40 – 1.00 p.m. Biostatistics for biomarker discovery and phenotype prediction Dr Etienne Thevenot French Alternative Energies and Atomic Energy Commission 1.00 – 1.20 p.m. Metabolomics-A great tool for generating hypothesis Dr Wang Yulan Wuhan Institute of Physics and Mathematics Chinese Academy of Sciences, China A20 A21 A22 A23 1.20 - 2.10 p.m. Prof Tang Huiru Wuhan Institute of Physics and Mathematics Chinese Academy of Sciences, China Lunch 17 MERLION METABOLOMICS WORKSHOP SINGAPORE 20 Nov 2014 (Thursday) Day 2 Abstract No. Session 6- Environmental Metabolomics Chaired by: Assoc Prof Sanjay Swarup and Dr Cyril Jousse A24 A25 2014 2.10 – 2.40 p.m. Keynote Speaker Model systems for the metabolomics study of environmental exposure Prof Robert Trengove Separation Science and Metabolomics Laboratory Murdoch University, Australia 2.40 – 3.00 p.m. Environmental metabolomics to understand resource partitioning strategies in algae Assoc Prof Sanjay Swarup NUS Environmental Research Institute National University of Singapore, Singapore A26 3.00 – 3.20 p.m. A27 3.20 – 3.40 p.m. Non targeted metabolomics for assessment of environmental exposure to contaminants and their biological effects: towards exposomics Dr Laurent Debrauwer MetaboHub-Toulouse French National Institute for Agronomy Research, France A study on Honeybee losses. First insight into environmental interactions. Dr Cyril Jousse University of Blaise Pascal, France 3.40 - 4.00 p.m. Tea Break Session 7 - Young researchers session Y9-16 4.00 - 6.00 p.m. 7.30 - 9.30 p.m. 8 oral presentations (15 min each) Presenters: Environmental • Mr Umashankar Shivshankar • Ms Gao Yan • Ms Zhang Wenlin • Dr Amit Rai • Dr Saravan Periasamy Food Science & Technology • Ms Nay Min Min Thaw Saw • Dr Aw Wanping • Dr Rajesh N. Patkar Chaired By: Dr Blandine Comte French National Research Institute for Agricultural Research, France Cocktail Reception and Dinner Banquet 18 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 21 Nov 2014 (Friday) Day 3 Abstract No. Session 8 - Environmental Metabolomics Chaired by: Prof Sam Li Fong Yau and Prof Robert Trengove A28 A29 A30 A31 A32 9.00 – 9.30 a.m. Keynote Speaker Comprehensive investigations of cellular metabolic networks using 13C-fluxomics and applications to systems microbiology and biotechnology Prof Jean Charles Portais MetaboHub-Toulouse University of Toulouse, France 9.30 – 9.50 a.m. Response of cloud microorganisms to atmospheric stresses: the case study of cold shock Dr Anne-Marie Delort University of Blaise Pascal, France 9.50 – 10.10 a.m. Analysis of leachates from gasification of solid wastes by metabolomics techniques Prof Sam Li Fong Yau NUS Environmental Research Institute National University of Singapore, Singapore 10.10 – 10.30 a.m. Methods for dealing with batch effects in metabolome data Dr Rohan Benjamin Hugh Williams Singapore Centre for Environmental Life Sciences Engineering National University of Singapore, Singapore 10.30 - 10.50 a.m. Metabolomics-driven strain improvement of higher alcohols in Escherichia coli 10.50 – 11.10 a.m. 11.10 a.m. - 12.30 p.m. 12.30 - 12.35 p.m. 12.35 - 12.40 p.m. Dr Sastia Prama Putri Department of Biotechnology Graduate School of Engineering Osaka University, Japan Tea break Panel Discussion on Metabolomics Research Directions Closing Remarks (France) Closing Remarks (Singapore) 19 Dr Fabien Jourdan MetaboHub-Toulouse French National Research Institute for Agricultural Research, France Assoc Prof Sanjay Swarup NUS Environmental Research Institute National University of Singapore, Singapore MERLION METABOLOMICS WORKSHOP SINGAPORE 12.40 - 1.30 p.m. 2014 Lunch Site 1: Agilent Factory in Yishun Industrial Estate 1.30 - 4.30 p.m. Technical Site visit Site 2: NUS Environmental Research Institute (NERI) Laboratory at T-Lab and CREATE. Singapore and French Discussion (By Invitation Only) Workshop End In Summary Item No. of Speakers Duration Keynotes 9 25 mins presentation and 5 mins Q&A Oral Presentations 23 15 mins presentation and 5 mins Q&A Young researchers presentations 16 10 mins presentation and 5 mins Q&A 20 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Young Researcher Award The “Young Researchers Award” recognizes and celebrates innovation in research as presented during the Merlion Metabolomics Workshop Young Researchers’ session. Selected members of the Workshop’s Scientific Committee have graciously agreed to serve as judges for the Award and their decisions are final. Two prizes shall be presented in 2014. Each prizewinner will receive a CRC Press book titled: “Mass Spectrometry-Based Metabolomics: A Practical Guide” (RRP USD140). Mass Spectrometry-Based Metabolomics: A Practical Guide is a simple, step-by-step reference for profiling metabolites in a target organism. The book summarizes all steps in metabolomics research, from experimental design to sample preparation, analytical procedures, and data analysis. Case studies are presented for easy understanding of the metabolomics workflow and its practical applications in different research fields. The book includes an in-house library and built-in software so that those new to the field can begin to analyze real data samples. In addition to being an excellent introductory text, the book also contains the latest advancements in this emerging field and can thus be a useful reference for metabolomics specialists. For more information: http://www.crcpress.com/product/isbn/9781482223767 Many thanks to Professor Eiichiro Fukusaki, Dr Sastia Putri And CRC Press for their sponsorship of this award. 21 MERLION METABOLOMICS WORKSHOP SINGAPORE Abstracts for Main Session 22 2014 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A1. Introducing metabolomics in Singapore Eric Chan Chun Yong 1, Sam Li Fong Yau 2, 5, Sanjay Swarup3 ,5 and Choon Nam Ong 4, 5 * 1 Department of Pharmacy Department of Chemistry 3 Department of Biological Sciences 4 School of Public Health 5 NUS Environmental Research Institute (NERI) National University of Singapore *E-mail: [email protected] (corresponding author) 2 Abstract Metabolomics is a burgeoning ‘omics’ field that leverages upon the established analytical chemistry technologies and bioinformatics tools for the study of small metabolites. In Singapore, a core group of scientists have been applying metabolomics during the past decade developing suitable technologies for metabolomics investigations and concurrently using this technology to solve real-world problems such as the discovery of novel metabolic signatures for the diagnosis of diseases, elucidation of effect of host-gut microbiota interactions on drug disposition, diet and disease, plant and microbial metabolomics, metabolic changes of exposure to environmental contaminants, and identification of biomarkers for early disease prevention and treatment. The team also working closely with researchers locally and internationally and the analytical platforms used including Nuclear Magnetic Resonance (NMR) and different Mass Spectrometry (HPLC-MS and GC-MS). The team also engaged commercial and industrial sectors to develop advanced technologies for metabolomic investigations. In this paper the authors will update some of their recent works on environmental and biomedical related metabolomics. The challenges and future of this field in Singapore will also be discussed. 23 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A2. MetaboHUB and RFMF: two tools at the service of metabolomics and fluxomics in France D. Rolin1*, F. Bellvert2, J. Bertrand-Michel2, C. Canlet2, R. Cole3, S. Colombié1, C. Deborde1, L. Debrauwer3, M. Ferrara4, L. Fouillen1, F. Jourdan2, C. Jousse4, C. Junot3, A. Moing1, JC. Portais2, E. Pujos-Guillot4, C. Sautot5, E. Thevenot3 1 MetaboHUB Bordeaux, INRA, CNRS, Université Bordeaux, F-33140 Villenave d'Ornon, France MetaboHUB Toulouse, CNRS, INRA, INSA, INSERM, Université Paul Sabatier, F-31000 Toulouse, France 3 MetaboHUB Saclay-Paris, CEA, University Paris VI, F-91190 Gif-sur-Yvette, France 4 MetaboHUB Clermont-Ferrand, INRA, Université Blaise Pascal, CNRS, F-63122 Saint-GenèsChampanelle, France 5 INRA Transfert, F-44300 Nantes, France *E-mail: [email protected] (corresponding author) 2 Abstract MetaboHUB and RFMF are two tools developed in France to serve the scientific community in Metabolomics and Fluxomics (M&F). The French Metabolomics and Fluxomics Network (RFMF, http://bit.ly/fr_metabolomics) is a scientific society which contributes to the development of M&F and more broadly to the overall analysis of metabolism in France through various actions: • Foster relationships between French researchers working in M&F (public and private sectors); • Promote and structure education and training in M&F; • Organize and support the organization of conferences in France and abroad; • Encourage the participation of young researchers to conferences through travel grants; • Allocate funds mission or prizes for accomplished work in M&F. In 2013, RFMF and the international Metabolomics Society formed an international affiliation to promote metabolomics. MetaboHUB project (http://www.metabohub.fr/en/) aims at creating a national infrastructure that will place France among the European leaders for advanced research services in M&F. MetaboHUB provides equipment and services to academic research and industrial partners in nutrition, health, agriculture, environment and biotechnology. MetaboHUB is the merging of 4 existing facilities (Bordeaux, Paris-Saclay, Toulouse and Clermont-Ferrand) into a unique infrastructure sharing common regulations and complementary M&F tools. Besides providing state-of-the-art services and support to national and international projects, a major ambition of MetaboHUB is to develop innovative tools and methods to address critical biological questions. MetaboHUB will provide custom solutions for (i) high-throughput, quantitative technologies for biochemical phenotyping of large sets of samples and for systems biology through standardization and combination of state–of-art technologies, (ii) identification of metabolites in human biofluids, plants, microorganisms and animal cell extracts, through the implementation and maintenance of centralized and open spectral repositories for metabolome annotations, (iii) large-scale flux profiling and sub-cellular fluxomics through integration of analytical data from multiple analytical devices, (iv) access to high-impact services to the national scientific community and industrial actors and, (v) attracting and training a new generation of scientists and students. 24 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A3. Metabolomics the way forward Royston Goodacre* School of Chemistry, and Manchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7DN, UK *E-mail: [email protected] (corresponding author) Abstract Metabolomics is a growing discipline that allows the analysis of the thousands of structural different small molecules found within a biological system. These metabolites can be measured using a variety of different analytical approaches including NMR spectroscopy or alternatively can be separated and quantified using gas or liquid chromatography coupled to mass spectrometry, after which the hope is that specific biomarkers can be discovered and linked to health, nutrition and disease [1]. This presentation will provide an overview of metabolomics and will give a flavor of how data are generated and analyzed. The role of metabolomics in nutrigenomics will also be discussed, as well as the concepts of the human being considered a superorganism [2] and all the complexities this generates that need to be overcome to understand human health and disease. During the presentation, I will also discuss the bioanalytical pathway [3] that we have developed as part of our human serum metabolome project (http://www.husermet.org/) that is need in order to negotiate the metabolite superhighway such that rigorously identified metabolites are discovered that can be used for understanding normal population behaviours and for disease stratification [4]. Finally I will give a very brief introduction to the Metabolomics Society. References 1. Goodacre, R. Vaidyanathan, S., Dunn, W.B., Harrigan, G.G. & Kell, D.B. (2004) Metabolomics by numbers - acquiring and understanding global metabolite data. Trends in Biotechnology 22, 245-252. 2. Goodacre, R. (2007) Metabolomics of a superorganism. Journal of Nutrition 137, 259S266S. 3. Dunn, W.B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., Brown, M., Knowles, J.D., Halsall, A., Haselden, J.N., Nicholls, A.W., Wilson, I.D., The Husermet consortium, Kell, D.B. & Goodacre, R. (2011) Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols 6, 1060-1083. 4. Dunn, W.B., Lin, W., Broadhurst, D., Begley, P., Brown, M., Zelena, E., Vaughan, A.A., Halsall, A., Harding, N., Knowles, J.D., Francis-McIntyre, S., Tseng, A., Ellis, D.I., O’Hagan, S., Aarons, G., Benjamin, B., Chew-Graham, S., Moseley, C., Potter, P., Winder, C.L., Potts, C., Thornton, P., McWhirter, C., Zubair, M., Pan, M., Burns, A., Cruickshank, J.K., Jayson, G.C., Purandare, N., Wu, F.C.W., Finn, J.D., Haselden, J.N., Nicholls, A.W., Wilson, I. D., Goodacre, R. & Kell, D.B. (2014) Molecular phenotyping of a UK population: defining the human serum metabolome. Metabolomics, in press 25 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A4. Metabolic network modelling and Food Toxicology Florence Vinson1, Ludovic Cottret2, Benjamin Merlet1, Yoann Gloaguen3, Clément Frainay1, Nathalie Poupin1, Daniel Zalko1, and Fabien Jourdan1* 1 INRA UMR1331 - Toxalim - MetaboHub, Toulouse, France INRA, Laboratoire des Interactions Plantes-Microorganismes (LIPM), UMR441, Castanet-Tolosan, F-31326, France 3 Glasgow Polyomics, University of Glasgow, 120 University Place, Glasgow G12 8TA * Speaker Mailing address: Fabien Jourdan, INRA UMR1331 TOXALIM-MetaboHub, 180 Chemin de Tournefeuille F31027 Toulouse Cedex 3, France *Email: [email protected] (corresponding author) 2 Abstract Metabolomics is a powerful tool to detect if there is a significant metabolic shift when an organism is exposed to chemical pollutants. To do so, lists of significantly modified metabolites, constituting metabolic fingerprints, are built using Nuclear Magnetic Resonance and/or Mass Spectrometry. It then allows classifying individuals based on the dose, frequency, and period of exposition to xenobiotics. Nevertheless these biomarkers cannot directly be used to decipher the mechanistic changes occurring in the organism. This interpretation is of outmost importance since endocrine disruptors like Bisphenol A may have long term impacts and lead to metabolic diseases. Our aim is to develop modelling approaches to help in understanding which parts of the metabolism are affected by chemical pollutants. To interpret global and untargeted data generated by metabolomics we need to consider metabolism at once and not to focus on individual metabolic pathways. To do so we use metabolic network modelling which gathers all metabolic reactions an organism can use for catabolic and anabolic processes. Human genome scale metabolic network recently published contains 7440 reactions and 2626 metabolites. The challenge is then to mine this network in order to find which cascades of reactions are responsible of biomarker concentration changes. We propose algorithms to build interpretable sub-networks from the whole network by extracting tenths of meaningful reactions. We will show that, based on sub-network comparisons, it is then possible to point out specific metabolic shifts induced by different doses of Bisphenol A. Finally, since reconstruction of metabolic networks is achieved using genomic information, it establishes a link between reactions, proteins and genes. Network can then be used as a relevant framework for polyomics studies. All these bioinformatics methods and metabolic networks are accessible through the MetExplore web server we are developing in the framework of MetaboHub. 26 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A5. Untargeted metabolomic approaches and data mining tools for marker discovery in nutrition B. Lyan1,2, F. Giacomoni1,2, C. Joly1,2, C. Migné1,2, J. F. Martin1,2, M. Pétéra1,2, J. Bourseau3, A. Napoli3, J. L. Sébédio2, B. Comte2, E. Pujos-Guillot 1,2 * 1 MetaboHUB Clermont-Ferrand, INRA, University Blaise Pascal, F-63122 Saint-GenèsChampanelle, France 2 INRA, UMR1019, UNH, F-63000 Clermont-Ferrand, France 3 LORIA, B.P. 239, F-54506 Vandoeuvre-lès-Nancy, France *E-mail: [email protected] (corresponding author) Abstract Metabolomics is a powerful tool in nutrition research for characterizing complex phenotypes and identifying biomarkers of specific physiological responses to different diets in the context of molecular pathways, physiology and health status. However, nutritional metabolomics is particularly challenging because of the low amplitude of effects induced by nutritional interventions or transitions, in comparison to the large variability in samples due to intra/inter-individual variations but also collection, storage and pre-treatment. Untargeted approaches using multiple analytical platforms are particularly of relevance to obtain a large analytical coverage of metabolites, which is essential because of the extreme chemical diversity of the endogenous and food metabolomes, ranging from sugars and lipids to polyphenols. However, in this context, important criteria for selection of an untargeted method are not only based on analytical coverage, but also on robustness of sample pre-treatment and on analytical reproducibility in terms of retention times, masses and signal intensities. We will illustrate the use of UPLC/HRMS and GC/MS based untargeted strategies for the study of food exposure, as well as for the characterization of metabolic phenotypes associated with development or prevention of chronic metabolic diseases. Indeed, the determination and the annotation of plasma and urine metabolic profiles constitute a powerful data-driven approach for the discovery of potential biomarkers of food intake and/or metabolic changes. However, irrespective of the type of analytical techniques used, these metabolomic strategies generate large amounts of data that need analyses and integration to extract the biologically meaningful information and enable the discovery of new knowledge. We will show that application of numerical and symbolic data mining methods based on clustering and pattern mining/formal concept analysis are particularly relevant to discover classes and sub-classes of individual phenotypes, as well as guide visualization and interpretation of data from nutritional metabolomics. 27 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A6. Lipidomic analysis of palm oil variability L. Fouillen1*, G-F. Ngando-Ebongue2, Y. Yuang1, V. Arondel1 and R. Lessire1 1 Laboratoire de Biogenèse Membranaire, UMR 5200, Université de Bordeaux/CNRS, CS20032, 33140 Villenave d’Ornon, France, 2 Centre de Recherches sur le Palmier à Huile de la Dibamba, Institut de Recherche Agricole pour le Développement, BP243 Douala, Cameroon. *E-mail: [email protected] (corresponding author) Abstract Palm oil is one of the most widely used vegetal oil in the human diet. Triacylglycerols (TAGs), the molecules composing the oil, contain three fatty acids esterified to a glycerol backbone. Palmitic and oleic acids (C16:0 and C18:1, respectively) represent about 85% of palm oil total fatty acids. It has been questioned whether the high level of saturated palmitic acid (4555%) might lead to increased coronary heart disease risk, which has generated suspicion in consumers for food that contains palm oil. A research program is currently being developed in Cameroon to investigate natural variability of fatty acid composition, with the long term aim of decreasing saturated fatty acid levels in palm oil through conventional breeding. Thus, we have analyzed oils from different genotypes commonly used for palm oil production for fatty acid content by GC-FID and for TAG composition by ESI-MSn. Differences between the various genotypes in the level of fatty acid distribution and content as well as in the TAGs composition have been observed. The variability for palmitic acid ranges from 15 to 45% and for oleic acid from 40 to 60%. The TAG composition showed more than 20 molecular TAG species, with high content of TAG 52:2, TAG 54:3 and TAG 54:4. Our results show that variability of fatty acid and TAG compositions exists, which will be useful to select genotypes more appropriate for human and industrial uses. Moreover, these data will allow us to study in more details the biochemical mechanisms responsible for this variability. 28 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A7. Novel Metabolites Involved in Pathogen-Host Interaction and Disease Control Naweed Naqvi*, Rajesh Patkar, Ziwei Qu Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, Singapore 117604 *Email: [email protected] (corresponding author) Abstract The rice-blast fungus, Magnaporthe oryzae, is a hemibiotroph that utilizes specialized infection-structures (the appressoria) to breach the host cuticle. Blast disease is a major deterrent to crop production and food security. Using mutant screening and chemical genomics, we identified Abc3 transporter function as being essential for pathogenesis in M. oryzae (1). We developed a novel platform technology to identify the Abc3 efflux Transport Substrate. ATS is a Digoxin-like steroidal glycoside, and essential for the host penetration step of Magnaporthe pathogenesis (2). ATS functions in regulating ion-homeostasis across appressorial plasma membrane and subsequently acts as an elicitor/effector of the host defense response during the biotrophic phase of rice blast. Interestingly, ATS (and digoxin) possesses a potent antifungal activity and is capable of inhibiting growth and pathogenesis in rice blast and Candida species (US Patent No. 20120003261). Recently, we identified an Antibiotic Biosynthesis Monooxygenase (ABM) as a Cytochrome P450-related function essential for Magnaporthe pathogenesis. ABM monooxygenase activity is directly involved in the biosynthesis of a novel metabolite that is secreted into the host cells to suppress the defense/resistance response therein. ABM plays an important role in the transition from biotrophy to necrotrophy in the blast disease cycle. Thus, the blast fungus directly interferes with hormonal signaling involved in pathogen recognition and defense system in rice. Our novel disease control strategies aim to disrupt such highly specific and intricate chemical communication at the host-pathogen interface. References 1. Sun, C.B., Suresh, A., Deng, Y.Z., Naqvi, N.I. (2006) A Multidrug Resistance Transporter in Magnaporthe is required for Host Penetration and for Survival during Oxidative Stress. The Plant Cell 18, 3686-3705. 2. Patkar, R., Xue, Y., Shui, G., Wenk, M., Naqvi, N.I. (2012) Abc3-mediated Efflux of an Endogenous Digoxin-like Steroidal Glycoside by Magnaporthe oryzae is Necessary for Host Invasion during Blast Disease. PLOS Pathogens 8(8), e1002888. 29 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A8. Mass spectrometry based metabolomics and lipidomics for the study rare diseases Benoit Colsch1, Samia Boudah1, 2 Etienne Thévenot3, Alexandre Seyer4, Simon Broudin4, Lydie Oliveira5, Florence Castelli1, Jean-Claude Tabet1,6, Christophe Junot1* 1 LEMM-CEA-Saclay, MetaboHUB-Paris, France; 2GlaxoSmithKline - Centre de recherche F.Hyafil,France; 3 Laboratory of Software Intensive Technologies (LIST), CEA-Saclay, MetaboHUBParis, France; 4 Profilomic SA, France ; 5CEA-iRCM-GReX, France ;6 Laboratoire de Chimie Structurale Organique et Biologique, IPCM/CNRS, Université Pierre et Marie Curie, France. *Email: [email protected] (corresponding author) Abstract The metabolome is the set of small molecular mass compounds found in biological media, and metabolomics, which refers to as the analysis of metabolome in a given biological condition, deals with the large scale detection and quantification of metabolites in biological media. It is a data driven and multidisciplinary approach combining analytical chemistry for data acquisition, and biostatistics, informatics and biochemistry for mining and interpretation of these data. Since the middle of the 2000’s, high resolution mass spectrometry (HRMS) is widely used in metabolomics, mainly because the detection and identification of metabolites are improved compared to low resolution instruments. Furthermore, thanks to their versatility, HRMS instruments are the most appropriate to achieve an optimal metabolome coverage, at the border of other omics fields such as lipidomics. Our team is involved in the development of tools based on liquid chromatography coupled to HRMS for the profiling of metabolites and lipids in various human biological media, such as plasma, cerebrospinal fluids and red blood cell extracts. We have also developed spectral databases for annotation of data sets and metabolite identification. The aim of this lecture will be to present HRMS based tools for metabolomics and lipidomics, and their relevance to the field of biomarker discovery for the diagnosis and follow-up of pathologies. This will be done through studies developed at the laboratory, mainly in the field of neurological diseases. 30 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A9. Lipidomics : a key tool for human health. Pauline LE FAOUDER 1, Aude DUPUY1, Fabien RIOLS1, Véronique ROQUES1, Nicolas CENAC 2, Thierry LEVADE 3, Hervé GUILLOU 4 , Bernard PAYRASTRE 5, Justine BERTRANDMICHEL1* 1 MetaboHUB-MetaToul-LIPIDOMIQUE, Inserm U1048, 1 Av. J Poulhes, 31 432 Toulouse, France CPTP, Inserm U1043,CHU Purpan, 31 024 Toulouse, France 3 CRCT, Inserm U1037, 1 Av. J Poulhes, 31 432 Toulouse, France 4 Toxalim UMR 1331, INRA , 180 Chemin de Tournefeuille, 31 027 Toulouse, France 5 I2MC, Inserm U1048, 1 Av. J Poulhes, 31 432 Toulouse, France *E-mail: [email protected] (corresponding author) 2 Abstract Lipidomics is an emerging field, where the structures, functions and dynamic changes of lipids in cells, tissues or body fluids are investigated. To this end, methods should be able to quantify numerous lipid molecular species. There are two main difficulties to face: the wide variability of the lipid structures and the large range of quantities of these molecules in biological samples. There are virtually no common features between sterols, fatty acids, sphingolipids and glycerolipids: despite their solubility in organic solvents, their polar and chromatographic properties are very different. A good example that illustrates the wide range of concentrations is phosphrylated sphingoid bases which are 1,000-10,000-fold less abundant than the major sphingolipid sphingomyelin. These 2 points cause real difficulties to quantify lipids. Global approaches are complex, as minor lipid species which can be key biomarkers in health are often erased from the data. So we chose targeted approaches to be able to quantify minor lipids. Sample preparation is a key point of lipidomics, usually a liquid-liquid extraction is performed which can be completed with a solid phase extraction to pre-concentrate the sample. Hydrolysis and derivatisation steps can be added if necessary. Then classical chromatographic tools are used to separate molecules with gaseous or liquid phase depending on the lipid family. While universal detectors like corona or flame ionization are still used, mass detectors are now most popular. We will present here the way to analyze sterols, fatty acids and their oxidized derivatives, glycerolipids and sphingolipids in biological samples, with a focus on some applications in cancer, inflammation 1 and metabolic diseases 2. References 1. Gobbetti T, Le Faouder P, Bertrand J, Dubourdeau M, Barocelli E, Cenac N, Vergnolle N. Polyunsaturated fatty acid metabolism signature in ischemia differs from reperfusion in mouse intestine.PLoS One. 2013 2. Ducheix S, Montagner A, Polizzi A, Lasserre F, Marmugi A, Bertrand-Michel J, Podechard N, Al Saati T, Chétiveaux M, Baron S, Boué J, Dietrich G, Mselli-Lakhal L, Costet P, Lobaccaro JM, Pineau T, Theodorou V, Postic C, Martin PG, Guillou H, Essential fatty acids deficiency promotes lipogenic gene expression and hepatic steatosis through the liver X receptor., J Hepatol. 2013 31 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A10. Urinary metabolomics revealed arsenic exposure-related metabolic alteration: a proof-of-concept study in a Chinese male cohort Jie Zhang*, Weipan Xu, Liangpo Liu, Qingyu Huang, Meiping Tian, Heqing Shen Key Lab of Urban Environment and Health, Institute of Urban Environment, Chinese Academy of Sciences, China *E-mail: [email protected] (corresponding author) Abstract The general population may be exposed to arsenic through various sources. Although targeted urinary biomonitoring of arsenic species provides the most accurate assessment of arsenic exposure, the arsenic-related metabolic perturbations are required to be evaluated. The objective of this proof-of-concept study is to investigate arsenic-induced phenotypic metabolome changes. Urinary arsenic species such as inorganic arsenic, methylarsonic acid, dimethylarsinic acid and arsenobetaine were quantified in a Chinese adult male cohort using high performance liquid chromatography (HPLC)-inductively coupled plasma-mass spectrometry. Urinary metabolomes were analyzed using HPLC-quadrupole time of flight mass spectrometry (qTOF-MS) and the arsenic-related metabolic changes were investigated by using a partial least squares discriminant analysis model. After adjustment for age, BMI, smoking and drinking, five potential biomarkers of testosterone, hippurate, acetyl-N-formyl-5-methoxykynurenamine (AFMK), serine and guanine were identified from 61 candidate metabolites, which indicated the exposure to arsenic induced endocrine disruption and oxidative stress in humans. Some dose-dependent trends have been also observed. The combined pattern of testosterone, hippurate, serine and guanine gave an AUC of 0.91 with sensitivity and specificity = 88% and 80%, respectively. We have demonstrated that metabolomics can be used to identify arsenic-related changes at the biomolecular level. The nature of the arsenic-related metabolic changes and trends may be used to further refine population based risk analysis of arsenic exposure. 32 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A11. A13. Noninvasive urinary metabonomic diagnosis of human bladder cancer Kishore Kumar Pasikanti1, Kesavan Esuvaranathan2, Yanjun Hong1, Paul C. Ho1, Ratha Mahendran2, Lata Raman Nee Mani2, Edmund Chiong2, and Eric Chun Yong Chan1* 1 Department of Pharmacy, Faculty of Science, National University of Singapore, 18 Science Drive 4, Singapore 117543 2 Department of Surgery, National University Health System, 5 Lower Kent Ridge Road, Singapore 119074. *E-mail: [email protected] (corresponding author) Abstract Cystoscopy is the gold standard clinical diagnosis of human bladder cancer (BC). As cystoscopy is expensive and invasive, it compromises patients’ compliance towards surveillance screening and challenges the detection of recurrent BC. Therefore, the development of a noninvasive method for the diagnosis and surveillance of BC and the elucidation of BC progression become pertinent. In this study, urine samples from 38 BC patients and 61 non-BC controls were subjected to urinary metabotyping using two dimensional gas chromatography time-of-flight mass spectrometry (GC×GC/TOFMS). Subsequent to data preprocessing and chemometric analysis, the orthogonal partial least squares discriminant analysis (OPLS-DA, R2X = 0.278, R2Y = 0.904 and Q2Y (cumulative) = 0.398) model was validated using permutation tests and receiver operating characteristic (ROC) analysis. Marker metabolites were further screened from the OPLS-DA model using statistical tests. GC×GC/TOFMS urinary metabotyping demonstrated 100% specificity and 71% sensitivity in detecting BC while 100% specificity and 46% sensitivity were observed via cytology. In addition, the model revealed 46 metabolites that characterize human BC. Among the perturbed metabolic pathways, our clinical finding on the alteration of the tryptophan-quinolinic metabolic axis in BC suggested the potential roles of kynurenine in the malignancy and therapy of BC. In conclusion, global urinary metabotyping holds potential for the noninvasive diagnosis and surveillance of BC in clinics. In addition, perturbed metabolic pathways gleaned from urinary metabotyping shed new and established insights on the biology of human BC. 33 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A12. Natural variation of lipidomes Markus R Wenk1,* 1 Department of Biochemistry, Yong Loo Lin School of Medicine, and Department of Biological Sciences, National University of Singapore *E-mail: [email protected] (corresponding author) Abstract Once viewed simply as a reservoir for carbon storage, lipids are no longer cast as bystanders in the drama of biological systems. The emerging field of lipidomics is driven by technology, most notably mass spectrometry, but also by complementary approaches for the detection and characterization of lipids and their biosynthetic enzymes in living cells (Wenk 2010 Cell 143(6):888-95). Our recent results show extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans (Chua et al 2013 Proc Natl Acad Sci U.S.A. 110(35):14468-73). I will also introduce a strategy for capture of phospho-monoester lipids. Using this enhanced workflow we identified novel forms of sphingosine-1-phosphates, in tissue from human, mouse and fruit fly, respectively. Understanding better the fundamentals of natural variation in lipidomes as well as specific recognition of individual lipid species are the scientific aims of SLING, the Singapore Lipidomics Incubator (http://lsi.nus.edu.sg/corp/research-programmes/sling/). This centre is a global magnet for collaborating parties in lipidomics – from academia and industry – delivering new technologies and intellectual capital. SLING organizes the international Singapore Lipid Symposium (ISLS), a major symposium in lipidomics research in Asia Pacific (http://www.lipidprofiles.com/index.php?id=82) and ‘i c lipid’, an intensive immersion course in mass spectrometry based lipidomics (http://www.lipidprofiles.com/index.php?id=139). 34 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A13. Application of metabolic fingerprinting for food quality assessments Eiichiro Fukusaki* Department of Biotechnology, Graduate School of Engineering, Osaka University, Japan *E-mail: [email protected] (corresponding author) Abstract Foods are generally multi-functional commodities which consist of multi components. It is almost impossible to perfectly elucidate the relationship between multi components and multi functions. Therefore, qualitative study of food function is very much difficult. A conventional reductionism would not be effective for food function study. Food sensory panel is very important for food quality evaluation. To organize good sensory test, skilled veterans must participate. But to keep enough number of skilled veteran with good condition is difficult. Alternative ways for food sensory test would be required. By the way, recently metabolomics is thought to be a promising tactics for further understanding genomic information. Metabolomics would be useful information in the integration of ome-information such as transcriptome and/or proteome. However, metabome itself can be also applicable without any ome-information. Particularly metabolic fingerprinting can be available in various application areas. Using metabolic fingerprinting technology, we can observe relationship between patients’ metabolome and their disease condition. It indicates usage of metabolome for early diagnosis system for some disease. The metabolic fingerprinting is also applicable to food materials. We can quantitatively guess several food functions including sensory value by means of metabolic fingerprinting. We conducted GC/MS and LC/MS to analyze compounds in food both qualitatively and quantitatively. The organized metabolome matrix was subjected to some multivariate analysis including PCA, PLS and OPLS. We have already reported metabolomics oriented sensory prediction of green tea, water melon, Japanese-sake, cheese, soy-sauce etc. I will introduce a principle of food metabolic fingerprinting. And some experimental example of them will be delivered. References 1. Putri SP, Yamamoto S, Tsugawa H, Fukusaki E., J Biosci Bioeng. 2013 Jun;115(6) 9-16. “Current metabolimics:Technological Advance”. 2. Putri SP, Nakayama Y, Matsuda F, Uchikata T, Kobayashi S, Matsubara A, Fukusaki E., J Biosci Bioeng. 2013 Jun;115(6):579-89. “Current metabolimics:Practical application”. 35 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A14. Metabolomics and systems biology in the context of food toxicology D. Zalko1*, C. Canlet2, N.J. Cabaton1, M. Tremblay-Franco2, M. Audebert1, N. Poupin1, E. Jamin2, R. Gautier2, L. Debrauwer2, F. Vinson1, F. Jourdan1. 1 MeX team, UMR 1331 Toxalim INRA-INPT, 31027 Toulouse cedex 3, France MetaboHUB Toulouse, CNRS, INRA, INSA, Université Paul Sabatier, 31077 Toulouse cedex 04, France *E-mail: [email protected] (corresponding author) 2 Abstract Novel questions in toxicology have emerged regarding chemicals that interfere with the homeostasis of living systems. Many man-made molecules present in the environment and the food-web are suspected EDC (Endocrine Disrupting Chemicals), and there's a growing awareness of the public regarding the risks associated with these bioactive substances. The evaluation of EDC sets new challenges for toxicologists as well as health risk assessment agencies, as illustrated by ongoing controversies on the “low-dose” effects of EDC1. Even for the best known model xeno-estrogens, mechanisms of low-dose metabolic modulation are only poorly understood, and the observation of non-monotonic dose-response curves (in vitro and in vivo)2,3 suggests a metabolic modulation based on multiple targets. Consequently, the debate has gradually evolved towards a major question currently discussed by toxicologists: should such effects be considered as adverse effects or are they simply an adaptive response with no health consequences? We previously demonstrated that the modulation produced by very low doses of EDC could be detected when exposure occurs during the perinatal period1. It is expected that the use of human metabolically competent cell lines, combined with high resolution spectral analyses, will allow, in a first step, to identify "metabolic fingerprints" of chemical pollutants. Then, based on a reconstruction of the metabolic network, and on comparative studies of closely related xenobitics (in terms of structure and/or effects), evidence for metabolic networks modulation should highlight major disruption routes of cellular metabolism. The use of non targeted approaches also opens the way to a better understanding of the mechanisms of toxicity of low doses of exposure, and of the effect of mixtures. Ultimately, in vitro and in vivo approaches should be combined for building novel strategies to characterize the danger associated with the exposure to major groups of chemicals. References 1. Vandenberg, L.N., Maffini, M.V., Sonnenschein, C., Rubin, B.S., Soto, A.M. (2009). Endocr Rev 30, (1), 75-95. 2. Cabaton, N.J., Canlet, C., Wadia, P.R., Tremblay-Franco, M., Gautier, R., Molina, J., Sonnenschein, C., Cravedi, J.P., Rubin, B.S., Soto, A.M., Zalko, D. (2013). Environ Health Perspect. 586-93. 3. Shioda, T., Rosenthal, N.F., Coser, K.R., Suto, M., Phatak, M., Medvedovic, M., Carey, V.J., Isselbacher, K.J. (2013). 36 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A15. Use of metabolomics for gene function discovery in a dwarf rice mutant Rengasamy Ramamoorthy1, Amit Rai1, Peter Benke2, S. Ramachandran3, Sanjay Swarup1,2, PRAKASH P. KUMAR1,2,3* 1 Department of Biological Sciences, Faculty of Science, National University of Singapore, Singapore 117543 2 NUS Environmental Research Institute, National University of Singapore 3 Temasek Life Sciences Laboratory, 1 Research Link, Republic of Singapore *E-mail: [email protected] (corresponding author) Abstract A dwarf mutant rice oscyp96B4 was identified from a phenotype screen of Ds insertion lines. The homozygous mutant plants also had reduced panicle size and seeding rates compared to the Nipponbare wild-type rice plants (WT). Molecular data showed that this mutant had a single copy Ds insertion in OSCYP96B4 gene. The gene function is unknown, but it is predicted to encode for a cytochrome P450 protein. Cytochrome P450s are monooxygenases, a family of membrane-bound heme-containing proteins that play various roles such as biosynthesis and metabolism of lipids, plant hormones and secondary metabolites. Quantitative Real-Time-PCR and Northern blot analysis showed that OsCYP96B4 was expressed in all the WT tissues tested, with high expression level in the reproductive organs and low level in the roots. Since functions of CYP96 gene family in rice are unknown, we employed a metabolomics approach to identify the probable function(s) of OSCYP96B4 gene. Total metabolite profiling and comparison was done in WT, mutant, dsRNAi, complementation line, and ectopic expression line of OSCYP96B4 gene. The LC-MS-based metabolomics analysis yielded ~8,000 ‘features’. The metabolomics-based plant clustering was consistent with observed phenotypes, and this has led to identification of several affected metabolic pathways in the mutant. Our data suggest that metabolomics is a useful tool to characterize mutants when the function of the affected gene is unknown. References 1. Ramamoorthy R, Jiang S-Y, Ramachandran S (2011). Oryza sativa Cytochrome P450 Family Member OsCYP96B4 Reduces Plant Height in a Transcript Dosage Dependent Manner. PLoS ONE 6(11):e28069 doi:10.1371/journal.pone.0028069 37 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A16. Nutritional metabolomics: an integrative understanding of metabolic disease development E. Pujos-Guillot1,2, M. Rambeau1, C. Joly1,2, B. Lyan1,2, J. F. Martin1,2, M. Pétéra1,2, A. Napoli3, J. L. Sébédio1, S. Czernichow4, M. Zins4, M. Goldberg4, B. Comte1* 1 INRA, UMR1019, UNH, F-63000 Clermont-Ferrand, France MetaboHUB Clermont-Ferrand, INRA, University Blaise Pascal, F-63122 Saint-GenèsChampanelle, France 3 LORIA, B.P. 239, F-54506 Vandoeuvre-lès-Nancy, France 4 UMS11 Cohortes en population, INSERM/Université Versailles Saint-Quentin, F-94807 Villejuif, France *E-mail: [email protected] (corresponding author) 2 Abstract Life course is a continuum of consecutive transitions with multifactor lifestyle changes potentially impacting well-being and health status. Among multiple factors, shifts in dietary pattern, food composition and processing, are causing nutritional transitions. In the context of deleterious behavior, these transitions led to energy imbalance with an increasing prevalence of non-communicable chronic diseases. The complexity of diseases brings challenges for an integrative understanding of the metabolic processes contributing to the development of these disorders. Metabolomics offers now the possibility of characterizing global alterations associated with disease conditions or nutritional exposition and transition, and of identifying early and/or predictive biomarkers of disease development. In this context, intervention studies are essential to analyse specific nutrition effects, to discover potential new biomarkers and identify their role in the pathophysiological mechanisms. Using this approach, we showed in normal and overweight volunteers submitted to two months of overfeeding, that metabolomics allows identifying trajectories that reveal early subtle metabolic dysregulations, not detected by classical biochemical approaches, linked to differences in metabolic plasticity. To obtain a more comprehensive picture of nutritional exposure as well as a better predictive capacity of (bio)markers, complementary study designs in cohorts are mandatory. Within the French Gazel cohort, we will show that multidimensional model building, consisting in integrating heterogeneous data (i.e. metabolic, metabolomic, clinical and functional but also socio-economic data and food patterns) will bring new tools to better predict metabolic syndrome development. These models will allow revealing determinants and modulators of changes, and evaluating the contribution of nutrition as a modifiable risk factor, in disease development. This work was performed through the technical and scientific support of MetaboHUB. It illustrates the constructive collaborations between the infrastructure and research units in order to tackle challenging scientific questions in nutrition. 38 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A17. A totally new way to measure the germination of grain using metabolomics O.A.H.Jones1*, M. Burke1 1 School of Applied Sciences, RMIT University, GPO Box 2476, Melbourne, VIC 3001, Australia *E-mail: [email protected] (corresponding author) Abstract The pre-harvest sprouting of grains frequently causes significant losses to grain growers worldwide. The problem can influence suitability of grain for processing and hence marketability and returns. It can occur throughout the grain growing regions of Australia and the result damage has adversely affected as much as 25 % of production in some seasons. Visual assessment of sprouting is ineffective because grains showing no external signs may still have sprouted sufficiently so that it is no longer possible for quality products to be made. As an example, it is known that Asian noodles are particularly sensitive to the presence of sprouted grain. Most procedures to determine whether a grain has germinated have been based around measuring the effects of particular enzymes that breakdown starch (α-amylase). This has been done either directly or indirectly, however, despite protracted efforts to develop enhanced ways to measure germination over the last 50 years, there are currently no highly accurate or reliable approaches available. The present work uses Infrared and Raman Spectrometry based metabolomic methods to measure germination of barley and wheat over 24 hours. Our results indicate that infrared spectrometry is not only able to tell if grain has germinated or not but also gives an indication of how far the germination process has progressed The technique therefore has the potential to form a simple dockside test that would allow segregation and minimising the comingling of sound and mildly sprouted grain. As a result grower returns can be maximised through ensuring customer satisfaction in the many markets where sprouting causes major problems with potentially large benefits to the grain industry. References 1. Jacobsen, J.V., Pearce, D.W., Poole, A.T., Pharis, R.P. and Mander L.N. (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiologia Plantarum 115, 428–441. 39 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A18. Metabolomic aspects of symbiosis and transgenomic interactions in Mammals H.R. Tang1,2*, Y.L. Wang2 1 Metabolomics and Systems Biology Laboratory, School of Life Science, Fudan University, Shanghai; 2 CAS Key Lab of Magnetic Resonance in Biological Systems, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan, China. *E-mail: [email protected] (corresponding author) Abstract All mammals are regarded as “superorganisms” consisting of two distinct but integrated parts including hosts themselves and symbiotic gut microbiota with the symbiosis established interactively through co-evolution and mutual selections. Mammals harbor trillions of microbes with thousands of microbial species in their gastrointestinal tract (GIT) which are also known as the gut microbiota, microparasites and microbiomes. In normal human GIT, there are more than one kilogram of microbes having ten times more cells than hosts. The gut microbiota co-develop with their hosts’ growth playing essential roles in many aspects of mammalian physiology and having profound effects on the hosts’ health. Therefore, microbiomes are regarded collectively as an “essential organ” or extended genomes, transcriptomes, proteomes and metabonomes for their mammalian hosts. The physiology and health of mammals in entirety have to be understood by taking into consideration of hosts, symbiotic microbes and their interactions. However, it is nontrivial to completely define the genomes of these microbiomes as for their hosts. Neither can their composition, transcriptomes and proteomes be defined in details since many species cannot be cultured ex vivo. Nevertheless, mammal and gut microbiota co-metabolism is essential for symbiosis or transgenomic interactions. This presentation will briefly introduce the importance of gut microbiota in mammalian physiology and pathophysiology. Then we will discuss some recent results on the mammalian metabolic interactions with their gut microbiota in terms of the environmental effects on human health with particular focus on the regulatory functions of gut microbiota in rodents and humans. Technical aspects will also be dealt with in the same context. References 1. 2. 3. 4. FPJ Martin, et al, Mol Systems Biol, 3, 112, 2007. M Li, et al, PNAS, 105, 2117, 2008. Y Tian, et al, J Proteome Res, 11,1397, 2012. Y Zhao, et al, J Proteome Res, 12,2987, 2013. 40 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A19. Metabolomic applications in bioprocessing and clinical research Y.S. Ho1,*, W.P.K. Chong1, C.C. Teo1, S.W. Chen1, S.Y. Mak1, E. Tan1, N. Basri1, E. Peh1, A.HM. Tan1, S. Xu1,2, D-Y. Lee1,3, T.H.K. Lim4, D.M.Y. Tan5, K-P. Lam1,2 1 Bioprocessing Technology Institute, A*STAR, 20 Biopolis Way, #06-01 Centros, Singapore 138668; 2Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117599; 3 Department of Chemical and Biomolecular Engineering, National University of Singapore, 4 Engineering Drive 4, Singapore 117576; 4Department of Pathology, Singapore General Hospital, 20 College Road, Academia, Level 7, Singapore 169856; 5National Cancer Centre, 11 Hospital Drive, Singapore 169610. *E-mail: [email protected] (corresponding author) Abstract Research at the Bioprocessing Technology Institute (BTI) encompasses various aspects of biopharmaceutical drugs production, including the engineering of novel cell lines and biomolecules, the optimisation of therapeutics production and purification in microbial and mammalian systems and the characterisation of bioprocesses and products using a suite of analytical and “-omics” technologies. Of these “-omics” technologies, metabolomics is considered the closest representation of the phenotype of the system, as metabolite levels are often seen as the final response of biological systems to genetic or environmental alterations. A liquid-chromatography mass spectrometry (LC-MS) based metabolomics platform was initially established at BTI, with the primary objective of guiding bioprocess development for the Chinese Hamster Ovary (CHO) cell system – the most widely used host for large-scale production of biopharmaceutical drugs. The platform led to the successful identification of key metabolites impacting cell growth, as well as novel cell engineering targets, subsequently resulting in a cell line with improved growth characteristics and increases in recombinant therapeutic protein yield. More recently, the metabolomics platform has also been extended to biomarker discovery and the elucidation of disease mechanisms in various clinical applications. To accommodate the profiling of clinical samples such as primary cells and biofluids that are highly limited in nature, additional platform capabilities aimed at increasing metabolite coverage and reducing the sample volume required for analysis were developed. This presentation will chronicle the evolvement of our platform, from its initial beginnings as a supporting “-omics” based technology for bioprocess optimisation, to its more recent applications in the clinical field. 41 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A20. Metabolomics driven the discovery of functional small molecules towards a diversity of biological innovations Leyu Yan, Wenna Nie, Xiaojuan Guo, Tianbing Guan, Qiang Li, Jiaoyan Chen, Haitao Lu* Laboratory for Functional Omics and Innovative Chinese Medicines, Chongqing University Innovative Drug Research Centre, Chongqing, 401331, China *E-mail: [email protected] (corresponding author) Abstract In vivo small molecules as necessary intermediates are involved in numerous critical metabolic pathways and biological processes associated with many essential biological functions and events1,2. In recent years, There is growing evidence to manifest that metabolomics has being emerged as a powerful tool for facilitating the discovery of functional small molecules which provides novel insights into a diversity of biological concerns involving disease diagnosis, therapeutic discovery and toxicology3,4. Yet, metabolomics holds promise in promoting widely biological innovations in a real world besides terming biomedical niche, which also yields a wealth of information about food and environmental questions concerning safety and toxicology. In this proposed presentation, brief introduction to metabolomics science shall be given to highlight its concept, protocol, manipulation and biological application for the first instance, and then a line of typical examples will be adopted to demonstrate how metabolomics combining with bioinformatic/genetic strategy was employed by our group to facilitate the discovery of functional small molecules whose pattern changes can efficiently dissect the complex mechanisms implicated in varied biological events via targeting the most affected metabolic pathways, such as disease progression, drug toxicity, pathogen virulence and drug discovery as well.. References 1. Nie, W.N., Yan, L.Y., Lee, Y. H., Guha, C., Kurland, I. J., and Lu, H.T. * (2014). Advanced mass spectrometry based multiple omics technologies to exploring the pathogenesis of hepatocellular carcinoma. Mass Spectrometry Reviews, Jun 2, doi: 10.1002/mas.21439, Epub ahead of print. (*Corresponding Author). 2. Yan, L. Y., Nie, W.N., Parker, T., Upton, Z., Lu, H. T. * (2013) Mass spectrometry based metabolomics facilitates the discovery of in vivo functional small-molecules with a diversity of biological contexts. Future Medicinal Chemistry, 5, 1953-1965. (*corresponding author). 3. Lv (or u), H. T*., Hung, C. S., and Henderson, J. P.* (2014): Metabolomic Analysis of Siderophore Cheater Mutants Reveals Metabolic Costs of Expression in Uropathogenic Escherichia coli. Journal of Proteome Research, 13, 1397-1404.(*Corresponding Author). 4. Lv (or u), H. T.* (2013): Mass spectrometry based metabolomics towards understanding of gene functions with a diversity of biological contexts, Mass Spectrometry Reviews, 32, 118-128. (*Corresponding Author). 42 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A21. Urine, metabolite profiling, its applications Eng Shi Ong Singapore University of Technology and Design, 20 Dover Drive, Singapore 138682. *E-mail: [email protected] (corresponding author) Abstract To evaluate and monitor body homeostasis and other processes in metabolism, urine has been an effective biofluids. Urine has a number of advantages over other biofluids for diagnostics purposes as a result of to its non-invasive nature. The formation of urine in the human body, various aspects of analysis of urinary metabolites including sampling and traditional approaches before presenting recent developments using metabolite profiling will be emphasized. Currently, the advancement of liquid chromatography tandem mass spectrometry (LC/MSMS), the establishment of standardized chemical fragmentation libraries, computing power, novel dataanalysis approaches with pattern recognition tools have made mass spectrometry-based metabolomics the latest sought-after technology. Metabolites present in the urine samples collected can vary widely and normalization for these effects will be presented. In addition, it will allow for the ability to dynamically catalogue and quantify small amount of cellular metabolites in urine samples. Case studies for colon cancer and others will be presented. It is noted that all these major advancement with mass spectrometry-based metabolomics will lead to new biomarkers and novel biochemical insights. References 1. Z Jiang, F Liu, ES Ong, SFY Li, Metabolic profile associated with glucose and cholesterol lowering effects of berberine in Sprague–Dawley rats. Metabolomics 2012, 8 (6), 10521068 2. Liu F, Gan PP, Wu H, Woo WS, Ong ES, Li SF. A combination of metabolomics and metallomics studies of urine and serum from hypercholesterolaemic rats after berberine injection. Anal Bioanal Chem. 2012 May;403(3):847-56. 43 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A22. Biostatistics for biomarker discovery and phenotype prediction Etienne A. Thévenot1,*, Natacha Lenuzza1, Aurélie Roux2, Samia Boudah2, and Christophe Junot2 1 Laboratory of Software Intensive Technologies (LIST) and 2 Drug metabolism research laboratory (LEMM), MetaboHUB Paris, CEA, 91191 Gif-sur-Yvette, France *E-mail: [email protected] (corresponding author) Abstract Biomarker discovery through metabolomics profiling shows promising results for the diagnostics of diseases such as cancer, Alzheimer's disease, or diabetes. Identification of an effective biomarker from thousands of raw LC-MS signals critically depends upon advanced statistical strategies for data processing, analysis, and the development of robust models. In particular, one of the major threats is bias resulting from confounding factors. To characterize the physiological variations of the human metabolome, we conducted a study of individual urine samples from more than 200 volunteers (CEA employees). Acquisitions in the positive and negative ionization modes were performed in a total of three batches. Preprocessing and annotation with public and in-house databases resulted in the structural full or partial identification of 400 metabolites. The variations of these metabolites associated to age, body mass index (BMI) and gender were analyzed with a three-step statistical methodology. First, analytical drifts (decrease of the sensitivity of the instrument over time during each run) and batch effects (offset from one run to the other) were corrected by using normalization approaches. Second, significance of the variation of each metabolite concentration associated to each factor (and their interactions) was determined by univariate hypothesis testing. Third, orthogonal partial least-squares (OPLS) algorithms were implemented to build and validate robust predictive multivariate models. Almost all gender-specific metabolites detected had higher concentrations in males compared to females, and included steroid hormones (e.g. testosterone) and the energy metabolism (lipids, dipeptides, and nucleosides). A decrease of metabolism with age was observed (steroid hormones, amino acids, nucleoside derivatives), with the exception of exogenous metabolites such as caffeine. Altogether these results provide a critical comparison of statistical approaches for biomarker discovery applied to the characterization of the human metabolome. 44 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A23. Metabolomics-A great tool for generating hypothesis Y. L. Wang*, H. D. Li, H. R. Tang Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, Wuhan Centre for Magnetic Resonance, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan, 430071, P. R. China *E-mail: [email protected] (corresponding author) Abstract Metabonomics is the science that studies dynamic alterations of metabolites in a cell, organ or entire organism1,2. The definition of the metabonomics was first given in 1999 as “the quantitative measurement of the time-related multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification”2. Technically, metabonomics investigations usually consist of the collection of metabolic profiles using nuclear magnetic resonance spectroscopy or mass spectrometry techniques and analysis of the collected data using appropriate multivariate statistical techniques. Metabonomics is a great tool for generating credible hypothesis. Recently, we have applied this technique to investigate interactions between host and hepatitis B virus (HBV). We characterize the metabolic features of host cells expressing the HBV virion using metabonomics, and then, identified important metabolic pathways related to the infection. Then we performed proteomics investigation to validate relevant metabolic pathways that were changed after HBV infection. Based on the metabolic and proteomics data, we found that HBV replication induces the promotions of central carbon metabolism, biosynthesis of nucleotides and total fatty acids; HBV up-regulates the biosynthesis of hexosamine and phosphatidylcholine through activating glutamine-fructose-6-phosphate amidotransferase 1 (GFAT1) and choline kinase α (CHKA), respectively. In order to validate the importance of GFAT1 and CHKA in the replication of HBV, we conducted further molecular biological assays on the system. The activations of GFAT1 and CHKA are known to contribute to carcinogenesis. Our results also indicate that HBV-induced hepatitis and fatty liver could be partially attributed to GFAT1 activated hexosamine biosynthesis. Furthermore, we demonstrate that GFAT1 and CHKA are two potential targets for treating HBV infection. This study provides new insights into the pathogenesis of HBV-induced diseases and the discovery of drug targets. We show in our work that metabonomics is a great tool for generating credible hypothesis. References 1. Nicholson, J. K., et al., 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29 (11), 1181-1189. 2. Tang, H. R., et al., Metabonomics: a revolution in progress. Prog Biochem Biophys, 2006, 33 (5), 401-414. 45 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A24. Model systems for the metabolomics study of environmental exposure Sarah Hayton1,2, Nicola Parkinson1,2, Stephen Harrison1,2, Stephanie Meilke1,2, Thomas Dignan1,2, Catherine Rawlinson1,2, Joel Gummer1,2,3, Ian Mullaney1,2, Garth Maker1,2, Robert Trengove1,3* 1 Separation Science & Metabolomics Laboratory, Murdoch University Veterinary and Life Sciences, Murdoch University 3 Metabolomics Australia, Western Australian Node *Email: [email protected] (corresponding author) 2 Abstract We are at the emergence of the capability to simultaneously measure residue and environmental analytes and profiles of primary and secondary metabolites. We can start to investigate from both directions or investigate "cause" and "effect" simultaneously. The life sciences, particularly proteomics and more recently metabolomics have been drivers in recent instrumental development, leading to faster chromatography, accurate mass and high resolution mass analyzers, and equally as important robust sample interfaces for minimal sample preparation. Alongside the hardware developments there have been considerable developments in software for mass spectral deconvolution, data visualisation, and mapping of analytes onto biochemical pathways to identify which pathways are modifed in a challenged sample vs a control sample. For instance, this allows the mechanisms of disease progression to be dissected and potential management strategies and potential therapeutic targets to be identified. These same instrumental developments have been utilised by the residue and environmental communities with the development of multiresidue methods that screen for in excess of 1000 analytes in a single analysis. Metabolomics seeks to identify changes in response to some stimulus, whilst residue and environmental analysis seeks to identify all possible residue and environmental "stimulus". This presentation will focus on, and address, the approach and the challenges to facilitate the use of both “targeted” and “untargeted methods”. In particular the different approaches required for GC/MS based vs LC/MS based metabolomics. Instrumental optimisation will be discussed in the context of combination of targeted compound analysis and metabolite profiling. 46 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A25. Environmental metabolomics to understand resource partitioning strategies in algae Swarup S1, 2, 3 *, Pan Di1, Amit Rai2, Umashankar S1, 2, 3, Vello V4, Kumar V1, Benke P1, Phang S M4, Williams RBH3 1 NUS Environmental Research Institute, National University of Singapore, Singapore 117411; 2 Metabolites Biology Lab, Department of Biological Sciences, National University of Singapore, Singapore 117543; 3 Singapore Centre for Environmental Life Sciences Engineering (SCELSE), Nanyang Technological University, Singapore 637551; 4 Institute of Ocean and Earth Sciences, University of Malaya *E-mail: [email protected] (corresponding author) Abstract Cellular resources in organisms are shared and reallocated dynamically to adapt to environmental pressures. We show the metabolic resource partitioning strategies used by algae in diverse environmental conditions, using two case studies (i) during harmful algal bloom for cell multiplication and toxin production, (ii) influence of habitat and growth on microalgae to guiding bioprocess development for biofuel production. (i) Microcystis aeruginosa PCC7806, a toxic model cyanobacterium strain, has been used in our laboratory to study the transcriptome and metabolome dynamics of cyanobacterium under different light and nitrogen conditions that trigger algal bloom. It was noticed that light influences the cellular processes mainly through transcriptional changes, while nitrogen’s effects are mainly seen at metabolome level. Under increased light conditions, both the microarray and metabolic profiling data suggested that combating oxidative stress, while maximizing growth is the preferred strategy for cyanobacteria. The most significant change in metabolites observed was for amino acids, and therefore, protein biosynthesis and amino acid metabolism were identified as pathways significantly altered during the shift from low to high microcystin conditions. High amino acid concentration at high nitrogen concentration is associated with low mcy gene expression. These results suggest that when there is more nitrogen available, the cellular strategy is to use more amino acids for protein synthesis, rather than microcystin biosynthesis. (ii) Algal growth and lipid production are interlinked, and play a critical role during biofuel production processes. Here, we used untargeted high-resolution mass spectrometry to understand the metabolic differences at exponential and stationary growth stages using 22 Chlorella strains collected from S.E. Asia. This analysis revealed strain-specific signatures of metabolic reprogramming strategies, and associations between physiochemical measures and metabolic pathways. These findings will aid the development of efficient bioprocess strategies to improve algal bio-products. 47 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A26. Non targeted metabolomics for assessment of environmental exposure to contaminants and their biological effects: towards exposomics L. Debrauwer1*, E. Jamin1, C. Canlet1, M. Tremblay-Franco1, N. Bonvallot2, C. Chevrier2, S. Cordier2, J.P. Cravedi3 1 MetaboHUB Toulouse, CNRS, INRA, INSA, Université Paul Sabatier, 31077 Toulouse cedex 04, France 2 INSERM UMR 1085 IRSET – EHESP, 35042 Rennes, France 3 MeX team, UMR 1331 Toxalim INRA-INPT, 31027 Toulouse cedex 3, France *E-mail: [email protected] (corresponding author) Abstract The use of pesticides and related environmental contaminations can lead to human exposure to various molecules, which could be responsible for adverse effects. However, human health risks associated with multi-exposure to complex mixtures are currently under-explored, and the following questions remain challenging: Can we assess the exposure to pesticides from biofluids in an untargeted way? What is the influence of exposures to multiple pesticides on the metabolome? What mechanistic pathways could be involved in the metabolic changes observed? To provide answers to these questions, both NMR and MS tools were developed, based on urine samples from 338 early pregnant women belonging to the PELAGIE cohort, which were classified in 3 groups according to the surface of land dedicated to cereal farming in their town of residence. For non-targeted exposure assessment, an LC-MS workflow was developed, integrating untargeted data acquisition, identification of pesticides metabolites (which structures could be unknown) using an upgradable list of metabolites to seek in urine samples, and statistical treatment of data. The groups of individuals could be separated according to their exposure to pesticides, on the basis of metabolites representing 2 fungicides (azoxystrobin, fenpropimorph) usually used on cereal cultures. The relevance and limitations of this innovative approach will be discussed. To evidence possible impacts of the exposure on global metabolism as revealed in urine, NMR-based analyses were performed. Global NMR fingerprints were treated using supervised multivariate statistics (PLS-DA, polytomous regressions) to discriminate the 3 groups of women, after restriction of our sample to subjects with 10-11 weeks amenorrhea and 20-30 body mass index (N=123) for reducing inter-individual variability. The most statistically significant changes (decreasing trend) were observed for glycine and citric, hippuric and lactic acids, indicating that a complex exposure to pesticides could induce glycine depletion (aminoacid involved in xenobiotics biotransformation) and disturbances of energy metabolism. 48 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A27. A study on Honeybee losses. First insight into environmental interactions. Céline DALLE1-3, Johan PANEK1, Angélique ABILA3, Hicham EL ALAOUI1, Marie LAGREE3, Mounir TRAÏKIA2,3, Michaël ROUSSEL1, Frédéric DELBAC1 & Cyril JOUSSE 2,3* 1 Clermont University, Blaise Pascal University, « Microorganisms: Genome and Environment » Laboratory, CNRS, UMR 6023, F-63000 CLERMONT-FERRAND, FRANCE 2 Clermont University, Blaise Pascal University, Institute of Chemistry of Clermont-Ferrand, CNRS, UMR 6296, F-63000 CLERMONT-FERRAND, FRANCE 3 Platform of Exploration of Metabolism, Blaise Pascal University, F-63000 CLERMONT-FERRAND, FRANCE Email: [email protected] (corresponding author) Abstract The honeybee (Apis mellifera) colony losses recorded during the last decades represent a major issue regarding agricultural yields, apiculture and biodiversity conservation. The origin of these losses is thought to be multicausal, with a strong emphasis on pesticides and parasites. Among the parasites, the microsporidian Nosema ceranae is the etiologic agent of the Nosemosis disease in A. mellifera. This obligate intracellular parasite develops in the epithelial cells of adult honeybee mid-gut. The infection can increase energetic demand in bees, suppress the immune response and may affect pheromone production. Such disturbances may have major consequences on bee metabolic pathways. Here we present an emerging project for that purpose. We have decided to perform metabolomics using different tissues, hemolymph (bee biofluid) and gut. Hemolymph is collected by centrifugation and gut by dissection. Samples can be analysed trough Metabolic Profiler (LC/MS-NMR) and/or HR-MAS. After data treatment and multivariate analyses (PCA, PLS), databases have to be screened on the highest relevant signals. Validation on putative metabolites is usually conducted using 2D-NMR and orbitrap HRMS experiments. Then, hyphenation techniques permit to increase metabolic coverage and a better chance for stress biomarker discovery. References 1. Aliferis KA, Copley T, Jabaji S (2012) Gas chromatography-mass spectrometry metabolite profiling of worker honey bee (Apis mellifera L.) hemolymph for the study of Nosema ceranae infection. J Insect Physiol 58, 1349–1359. 2. Dussaubat C, Maisonnasse A, Crauser D, Beslay D, Costagliola G, Soubeyrand S, Kretzchmar A, Le Conte Y (2013) Flight behavior and pheromone changes associated to Nosema ceranae infection of honey bee workers (Apis mellifera) in field conditions, J Invertebr Pathol 113, 42-51. 3. Goblirsch M, Huang ZY & Spivak M (2013). Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS ONE 8, e58165. 49 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A28. Comprehensive investigations of cellular metabolic networks using 13 C-fluxomics and applications to systems microbiology and biotechnology JC. Portais1,2*, L. Peyriga1, F. Bellvert1, S. Sokol1, S. Heux2, F. Letisse2, L. Gales1, H. KulykBarbier1, E. Cahoreau1. 1 MetaboHUB Toulouse, CNRS, INRA, INSA, INSERM, Université Paul Sabatier, 31077 Toulouse cedex 04, France 2 Team MetaSys, Laboratoire d’Ingénierie des Systèmes Biologiques et des Procédés, CNRS, INRA, INSA, F-31076 Toulouse, France *E-mail: [email protected] (corresponding author) Abstract Metabolism is a basic cellular function that sustains survival, growth and adaptation of living organisms. At the cellular level, metabolism is organized as a network, i.e. a complex set of biochemical reactions that are tightly interconnected. Currently, the general – design – principles of organization, functioning and adaptation of metabolic networks are not well understood. Such knowledge is critical to get better understanding of the role of metabolism in the adaptation of organisms to their environment or to develop efficient and cost-effective biotechnological processes. In the recent years, we have developed a wide range of approaches (metabolomics, fluxomics) to provide detailed measurement of the response of metabolic networks to changes in the genome or environment. These methods can be combined with other omics tools (e.g. transcriptomics, proteomics, etc.) and to metabolic modelling to provide systems biology approaches for in-depth, comprehensive understanding of the organization, functioning and adaptation of cellular metabolism. This understanding is used to get efficient control and optimization of the metabolism of industrially-relevant microorganisms (e.g. E. coli, yeast). The use of systems-level studies makes it possible to comprehensively understand not only the impact of pathway engineering within the context of the entire host metabolism, but also to diagnose stresses due to product synthesis or to extreme environmental conditions, thereby providing the rationale for efficient and cost-effective optimization of production systems (knowledge-based metabolic optimization). In this lecture, the benefit and limits of these approaches will be discussed, with a particular emphasis on 13C-fluxomics. 50 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A29. Response of cloud microorganisms to atmospheric stresses: the case study of cold shock Anne-Marie Delort1,2,3*, Cyril Jousse1,3, Céline Dalle1, Isabelle Canet1, Marie Lagrée3, Mounir Traïkia1,3 , Bernard Lyan4 , Martine Sancelme1, Pierre Amato1,2 . 1 Clermont Université, Université Blaise Pascal, Institut de Chimie de Clermont-Ferrand, BP 10448, F-63000 Clermont-Ferrand , France 2 CNRS, UMR 6296, ICCF, BP 80026, F-63171 Aubière, France 3 MetaboHUB, PFEM, Université Blaise Pascal, BP 80026, F-63171 Aubière, France 4 MetaboHUB, PFEM, INRA-Clermont-Theix, Saint-Genes Champanelle, France *E-mail: [email protected] (corresponding author) Abstract Microorganisms have been known for a long time as important actors in aquatic and terrestrial ecosystems. Their discovery is rather recent in the atmosphere and more particularly in clouds (1). Our team has isolated around 700 microbial strains from cloud waters at the puy de Dôme summit during the last ten years, among them, Pseudomonas is one of the major encountered genus (2). Atmosphere is a really specific and versatile compartment on earth where microorganisms are facing various stresses including dryness, pH, UV, cold shocks…etc. Then, a key question is to understand how microorganisms can resist to such stresses and survive under such harsh conditions. In this work, we have selected a Pseudomonas syringae strain, isolated from cloud waters to study the influence of temperature and identify cold shock biomarkers through metabolomics experiments. This strain was incubated at 5°C and 17°C, these temperatures correspond respectively to the annual and the summer temperature means. Bacterial extracts were analyzed using a LC/MSNMR facility (Metabolic Profiler®), and multivariate analyses (PCA, PLS-DA). We highlighted different groups of relevant biomarkers. Some of them have been already described for cold shock stress: they include trehalose, betaïne, carnitine (cryoprotection ), organic acids, ATP, sugars (energetic metabolism ), insaturated lipids (membrane fluidity) and gluthatione (oxidative stress metabolism). Surprisingly, we also found short peptides described for the first time as cold shock biomarkers. One of these peptides was identified as a virulence factor (Tabtoxin). References 1. Delort et al., Atmos. Res., 2010, 98, 249-260. 2. Vaitilingom et al., Atmos. Environ., 2012, 56, 88-100. 51 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A30. Analysis of leachates from gasification of solid wastes by metabolomics techniques S. N. Lee1,3, W. L. Zhang1,3, L. Rong2,3, T. Maneerung3, C. H. Wang2,3, K. G. Neoh2,3, and S. F. Y. Li1,3* 1 Department of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543 Department of Chemical and Biomolecular Engineering, National University of Singapore, 4 Engineering Drive 4, Singapore 117576 3 NUS Environmental Research Institute, National University of Singapore, 5A Engineering Drive 1, Singapore 117411 *E-mail: [email protected] (corresponding author) 2 Abstract Leachate samples from fly ash and bottom ash obtained by gasification of solid wastes were analyzed by non-targeted screening using liquid chromatographyquadrupole-time of flight-mass spectrometry (LC-QTOF-MS). The results were used to determine which organic compounds could contribute to the toxicity of the leachates of solid waste gasification. Subsequently, the effects of the leachate on mortility and immobility of Daphnia mangna were evaluated as a method for monitoring water quality, and as a screening method for toxicity assessment of solid waste re-utilization. 52 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A31. Methods for dealing with batch effects in metabolome data S. Umashankar1, S. Swarup1,2, R. B. H. Williams2,* 1 Department of Biological Sciences, National University of Singapore, Singapore 117411; Singapore Centre for Environmental Life Sciences Engineering (SCELSE), National University of Singapore, Singapore, 117456 *E-mail: [email protected] (corresponding author) 2 Abstract High-throughput mass spectrometry based experiments generate large and complex data. Combining data from such experiments performed over time (weeks/months) or in different batches present numerous challenges. Due to the massively parallel nature of metabolomics data, such differences in experimental condition can induce a structure into the data that is of non-biological origin. These effects are often overlooked in current pipelines, but can easily contribute to artefactual signals being attributed to biological . Using untargeted metabolite profile data from 22 Chlorella strains, compared over two growth stages and run in four batches, we show removal of run-day effect, using a filtering procedure based on the singular value decomposition, that demonstrably preserves biological signal in the experiment, as measured by inter-strain variation. Our approach will be broadly applicable in metabolomics analysis, and highlights the importance of carefully dissecting the possible source into batch variation at the both the design and analysis stages of a study. 53 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 A32. Metabolomics-driven strain improvement of higher alcohols in Escherichia coli Sastia Prama Putri1, Naoki Kawase1, Shingo Noguchi1, Claire Shen2, James Liao2, Takeshi Bamba1, Eiichiro Fukusaki1* 1 Department of Biotechnology, Graduate School of Engineering, Osaka University, 2-1Yamadaoka, Suita, Osaka 565-0871, Japan; 2 Department of Chemical and Biomolecular Engineering, University of California, Los Angeles, 5531 Boelter Hall, 420 Westwood Plaza, Los Angeles, CA 90095, USA *E-mail: [email protected] (corresponding author) Abstract The concerted effort for bioproduction of higher alcohols to replace fossil fuels has translated into successful production of higher alcohols based on biochemical knowledge and stochiometric arguments. To date, the selection of targets for strain improvement has been based at its best on expert knowledge, but to a large extent also on ‘educated guesses’ and ‘trials and errors’. Metabolomics, the global quantitative assessment of metabolites in a biological system in combination with multivariate data analysis (MVDA) tools, allow the replacement of current empirical approaches by a scientific approach towards the selection and ranking of targets. In addition, metabolomics provides experimental data for identifying unknown and under-appreciated reactions in the metabolic network. Previously, Escherichia coli strains capable of producing 1-propanol from glucose had been constructed by incorporating synergy as a design principle of strain engineering. 1-propanol production was achieved via 2-ketobutyrate (2KB) from two pathways, native threonine pathway and non-native citramalate pathway from Methanococcus jannaschii. The strain possessing two pathways resulted in a higher titer of 1propanol (2.5 g/L/day), more than 3-folds compared with the production from a single pathway (0.8 g/L/day). Interestingly, SYN12 strain bearing overexpression of both pathways and SYN3 strain bearing overexpression of only citramalate pathway with a native threonine pathway had the same production performance. Here, we demonstrate the utility of metabolomics analysis to validate the importance of synergy for 1-propanol production, to effectively identify 2KB accumulation as the bottleneck step of 1-propanol production in strain SYN12, and to uncover the underlying reason of this bottleneck. 54 MERLION METABOLOMICS WORKSHOP SINGAPORE Abstracts for Young Researchers Session 55 2014 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y1. A comparative urine metabolic profiling of tuberculosis patients and healthy individuals Mrinal Kumar Das1*, Subasa Chandra Bishwal1, Aleena Das1, George M. Varghese2, Bhaswati Pandit3, Ranjan Nanda1 1 Immunology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), Aruna Asaf Ali Marg, New Delhi, India 2 Department of Medicine, Christian Medical College, Vellore, Tamilnadu, India 3 National Institute of Biomedical Genomics, Kalyani, West Bengal, India *E-mail: [email protected] (Presenter) Abstract: Tuberculosis (TB) is a curable disease and timely diagnosis could save many precious lives. However, most common TB diagnostic methods e.g. Sputum microscopy have limitations like low sensitivity and other logistic issues. Moreover, metabolite markers from non-invasively collected biofluid like urine hold promise of an easy to use, cost-effective and indextrous method for TB diagnosis. Research from our lab has demonstrated presence of distinct urine volatile signature in TB patients (Bandey et al., 2011). Here we are reporting deregulated non-volatile polar molecules in urine of TB patients. Urine samples (n=30; TB/Healthy: 17/13) were profiled using gas chromatography and mass spectrometry (GC-MS) preceded by trimethylsilyl derivatization. Raw data were preprocessed using ChromaTOF. The final metadata with the aligned molecules present in >50% of at least one class were exported as a .csv file for further analysis. Important molecules were identified using multi-variate analysis reported earlier by Das et al 2014. Pathway enrichment assay was carried out using a sub group of these selected molecules. Preprocessing of the raw data file resulted a 30x233 matrix and out of which 182 variables tentatively identified based on library search. A set of 21 molecules with VIP score more than 1.5 were selected as important molecules. Calculated area under curve (AUC) of receiver operating characteristic curve (ROC) was 0.96. Pathway enrichment showed altered lipid and amino acid metabolism in TB patients and corroborates findingof Zhou et al (2013). Our findings provide evidence that deregulated urine metabolites of TB patients may be useful as an alternate solution to develop new and easy to use TB diagnostic assay. References: 1. Banday, K.M., (2011) Anal. Chemistry 83, 5526–5534. 2. Das, M.K., (2014) Anal. Chemistry 86, 1229−1237. 3. Zhou, A., (2013) J. Prot. Research 12, 4642−4649 56 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y2. Metabolomics study of a mouse model with influenza virus induced lung injury Liang Cui1, Dahai Zheng1, Yadunanda Kumar1, Yie Hou Lee1, Wanxing Eugene Ho2, Jian Zhu Chen1,3, Choon Nam Ong2,4*, Steven R. Tannenbaum1,5* 1 Interdisciplinary Research Group in Infectious Diseases, Singapore-MIT Alliance for Research & Technology (SMART), Singapore 2 Saw Swee Hock School of Public Health, National University of Singapore, Singapore 3 Departments of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts, United States of America 4 NUS Environment Research Institute, Singapore 5 Departments of Biological Engineering and Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, United States of America *E-mail: [email protected] (Presenter) Abstract Influenza virus infection (IVI) can cause lung injury, which may progress to respiratory failure and fatal outcome. At present, the molecular interactions between host and influenza virus and corresponding metabolic processes involved in the viral pathogenesis are poorly-elucidated. We conducted a mass spectrometry-based metabolomics analysis on serum, bronchoalveolar lavage fluid (BALF) and lung tissue from a murine IVI model of lung damage and repair. To capture the dynamic metabolic responses to IVI and discover reliable differential metabolites related to influenza pathophysiology, biological samples were collected at 0, 6, 10, 14, 21 and 28 days post IVI, representing infection stages which correspond to ‘maximal viremia’, ‘peak body weight loss’, ‘maximal inflammatory lung damage’, and ‘recovery phase’ of the infection. Significant metabolome changes with over 100 identified differential metabolites were found upon IVI, covering a variety of metabolite classes. Meanwhile, distinctive metabolite profiles were observed in serum, BALF and lung, and certain metabolic changes were lungspecific, indicating the necessity of parallel metabolic profiling of both biofluids and lung tissues. Amongst various altered metabolic pathways, amino-sugar and nucleotide-sugar metabolism, purine and pyrimidine metabolism, sphingolipid metabolism, pantothenate and CoA biosynthesis, and tryptophan-kynurenine metabolism were identified to be involved in lung injury and recovery. These results advance current molecular understanding on host and influenza virus interactions, and provide mechanistic insights into influenza virus-induced lung injury and repair process. References 1. Kumar, Y., Cui, L., Limmon, G.V., Liang, L., Engelward, B.P., et al. (2014) Molecular analysis of serum and bronchoalveolar lavage in a mouse model of influenza reveals markers of disease severity that can be clinically useful in humans. PLoS ONE 9: e86912. 2. Cui L, Lee YH, Kumar Y, Xu F, Lu K, et al. (2013) Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl Trop Dis 7: e2373. 57 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y3. Metabolomics Studies of Urine and Plasma of Patients with Chronic Kidney Disease by LC-MS F. Liu1,2, X. Yin1, J. T. M. Ong1, J. B. W. Teo3, S. F. Y. Li1,2* 1 NUS Environmental Research Institute, 5A Engineering Drive 1, T-Lab Building, National University of Singapore, Singapore 117411; 2 organization and address , Calibri 11 pt 2 Department of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543 3 Department of University Medicine Cluster, National University Hospital, 5 Lower Kent Ridge Road, Singapore 119074 *E-mail: [email protected] (corresponding author) Abstract Chronic kidney disease (CKD) is a condition characterized by the progressive loss in renal function over a period of months or years which eventually leads to End Stage Renal Disease. Currently diagnostic biomarkers is not sensitive, therefore the aim of this study was to (i) characterize the wide-ranging metabolic changes occurring in CKD through non-targeted metabolomics; (ii) determine if there is any difference in the pathological manifestation of CKD in the non-diabetic group as compared to the diabetic group; (iii) identify metabolites for use as biomarkers of kidney function. To understand the system’s metabolic changes associated with CKD, we analysed the metabolomics changes of plasma and urine samples of patients with nondiabetic and diabetic CKD using LC-QTOF-MS. Statistical analyses such as OPLS-DA models were used to compare the metabolomics profiles and select the metabolites that showed significant change between diseased and control groups. The results presented here reveal alterations in uremic toxins, amino acids, glucose and ceramides in the plasma of CKD patients and decreased urinary excretion of bile acids, carnitines and vitamin E metabolites. These metabolomics alterations collectively indicate a state of oxidative stress, inflammation and acidosis in the pathophysiology of CKD. References 1. Weiss, H. R.; Kim, K. (2012) Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol 8, 22-33. 58 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y4. Metabolomic Studies of Acute Coronary Syndrome Patients using LCMS X.J. Yin1, 2*, F.Y.S. Li1,2,3 1 NUS Environmental Research Institute, 5A Engineering Drive 1, T-Lab Building, National University of Singapore, Singapore 117411; 2 Department of Chemistry, Block S5-02-03, 3 Science Drive 3, National University of Singapore, Singapore 117543 3 Shenzhen Engineering Laboratory for Eco-efficient Polysilicate Materials, School of Environment and Energy, Peking University Shenzhen Graduate School, Shenzhen, P.R.C. 518055 *E-mail: [email protected] (corresponding author) Abstract Discovery of new biomarkers is critical for early diagnosis of acute coronary syndrome (ACS). Recent advances in metabolomic technologies have drastically enhanced the possibility of improving the knowledge of its physiopathology through the identification of the altered metabolic pathways. In this study, peripheral plasma samples from 25 ST segment elevation ACS patients and non-ACS patients were collected before and 24 hours after the angiography. Analysis by liquid chromatography–mass spectrometry (LC–MS) and orthogonal partial least squares (OPLS) permitted the identification of many metabolites with statistical differences (p<0.05) between experimental groups. Additionally, validation by LC–MS permitted us to identify a potential panel of biomarkers formed by various amino acids and glucose metabolism. This panel of biomarkers reflects the oxidative stress and the hypoxic state that suffers the myocardial cells and consequently constitutes a metabolomic signature of the atherogenesis process that could be used for early diagnosis of ACS. References 1. Roger, V. L., Go, A.S., Lloyd-Jones. D.M., Adams. R.J., Berry. J.D., Brown, T.M., Carnethon, M.R., Dai, S., de Simone, G., Ford, E.S., Fox, C.S., Fullerton, H.J., Gillespie, C., Greenlund, K.J., Hailpern, S.M., Heit, J.A., Ho, P.M., Howard, V.J., Kissela, B.M., Kittner, S.J., Lackland, D.T., Lichtman, J.H., Lisabeth, L.D., Makuc, D.M., Marcus, G.M., Marelli, A., Matchar, D.B., McDermott, M.M., Meigs, J.B., Moy, C.S., Mozaffarian, D., Mussolino, M.E., Nichol, G., Paynter, N.P., Rosamond, W.D., Sorlie, P.D., Stafford, R.S., Turan, T.N., Turner, M.B., Wong, N.D., Wylie-Rosett, J. and American Heart Association Statistics Committee and Stroke Statistics Subcommittee. (2011). Heart disease and stroke statistics-2011 update: A report from the American Heart Association. Circulation, 123(4), e18–e209 2. Bassand, J. P., Hamm, C. W., Ardissino, D., Boersma, E., Budaj, A., Fernández-Avilés, F., Fox, K.A., Hasdai, D., Ohman, E.M., Wallentin, L. and Wijns, W. (2007). Guidelines for the diagnosis and treatment of non-ST-segment elevation acute coronary syndromes. European Heart Journal, 28, 1598–1660 59 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y5. LC-MS based metabolomics approach to study biomarkers of aging and chronic kidney disease E. Simsek1, L. F. Y. Sam2,* 1,2 NUS Environmental Research Institute, 5A Engineering Drive 1, T-Lab Building, National University of Singapore, Singapore 117411 *E-mail: [email protected] (corresponding author) Abstract Aging is a complex process involving the accumulation of diverse detrimental changes in biological systems gradually so significant changes in cellular metabolism are produced throughout aging. Normal aging process also often accompanies various diseases such as Alzheimer’s and Parkinson’s disease, diabetes and kidney disease. To investigate metabolic changes involved in aging process and chronic kidney disease (CVD), human plasma samples of healthy people and CKD patients of various ages, namely 40s, 50s, 60s and 70s, obtained from a local hospital in Singapore. The samples were analyzed using liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Multivariate statistical analysis of the data using principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) revealed separation between healthy versus CKD subjects and different age groups among healthy subjects. Metabolic features responsible for the separation between groups were chosen using variable importance for the projection (VIP) plot and searched against accurate mass metabolite libraries of METLIN metabolite database and Human Matabolome Database (HMDB) for identification. The study showed that it is possible to study biomarkers and related biological mechanisms related to aging and CKD with a mass spectroscopy based metabolomics approach. References 1. Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–83 (2011). 2. Mishur, R. J. & Rea, S. L. Applications of mass spectrometry to metabolomics and metabonomics: detection of biomarkers of aging and of age-related diseases. Mass Spectrom. Rev. 31, 70–95 (2012). 3. Zhu, Z.-J. et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 8, 451– 60 (2013). 60 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y6. Comprehensive metaoblomics study of the acute inflammatory response in a porcine model of combat trauma injury A.K.C. Laserna1, G. Fang1, J. Wu2, Y. Lai2, R. Ganapathy 2, S.M. Moochhala2, S.F.Y. Li1* 1 Department of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543; 2 Defence Medical & Environmental Research Institute, DSO National Laboratories, 27 Medical Drive, Singapore 117510 *E-mail: [email protected] (corresponding author) Abstract Severe injuries, such as trauma injury, are among the leading causes of morbidity and mortality worldwide.[1, 2] Injuries sustained in combat scenarios are even more dangerous given that the wounding agents are intentionally more lethal and the resources available for treatment are limited. Accompanying such severe injuries is the activation of the body’s immunologic and inflammatory response. Normally, they are meant to facilitate healing and protect the body from further damage. However, once they proceed uncontrolled, systemic inflammatory response syndrome (SIRS) may ensue subsequently leading to damage of multiple organs, and possibly death. The identification of biomarkers that can serve as indicators of the extent and progression of the inflammatory response and the onset of organ failure can help in determining the appropriate mode of intervention in the combat field, thereby improving patient outcome. In this study, we have taken an untargeted metabolomic approach using the complementary techniques of NMR and LC-MS to see the metabolic perturbations occurring in a simulation of combat trauma injury using a pig (porcine) model. Based on an established complex combat trauma injury model [3], pigs were subjected to femur fracture and soft-tissue injury with 60% blood loss, followed by a 30-minute shock period, and induced hypothermia. Plasma samples before and after injury, as well as sham samples, were analyzed to evaluate the changes in the metabolic profiles of the pigs. Significant changes in several metabolites were observed in response to the trauma injury. Correlation analysis with cytokines and some established protein markers revealed potential biomarkers of systemic inflammatory response syndrome (SIRS) and organ-specific injury. References 1. Global burden of disease: 2004 update. [Accessed 16 June 2011]; Available from: http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.h tml 2. The global burden of injuries. American Journal of Public Health, 2000. 90(4): p. 523-526. 3. Cho, S.D., et al., Reproducibility of An Animal Model Simulating Complex Combat-Related Injury in A Multiple-Institution Format. Shock, 2009. 31(1): p. 87-96. 61 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y7. Development and Applications of CE-QqQMS-based Metabolomics for Cancer Research K. Kami1,*, K. Sasaki1, T. Fujimori1, H. Yamamoto1, Y. Ohashi1 1 Human Metabolome Technologies, Inc., Mizukami 246-2, Kakuganji, Tsuruoka, Yamagata 9970052, Japan *E-mail: [email protected] (corresponding author) Abstract Most metabolites comprising primary energy metabolism such as glycolysis, pentose phosphate pathway, and the TCA cycle are anionic compounds; however, concentrations of most these metabolites in typical mammalian cells and tissues are often low and difficult to be measured by conventional metabolomics platforms. Here, we developed a capillary electrophoresis-triple quadrupole mass spectrometry (CE-QqQMS)-based metabolomics technology, which improved detection sensitivity of 63 anions up to 291-folds compared to a conventional time-of-flight mass spectrometry-based analysis. The method was then applied to profile metabolomic changes of cell lines treated by a Mek inhibitor, which significantly downregulated glycolysis and nucleotide synthesis in addition to oxidative stress and amino acid imbalance. The CE-QqQMS was also used to elucidate drug toxicity against cancer metabolism in EGFR sensitive/insensitive cell ilnes1. Paired non-tumor and tumor tissues of lung, prostate, and gastric cancer patients were also characterized. The results not only distinguished tumor from non-tumor profiles but also delineated within-tumor differences associated with their tissue types or differentiation status2; considerably high glycolytic intermediate, nucleotide, and glutathione levels in differentiated (DT) tumor tissues and rather low energy and redox statuses in undifferentiated (UT) ones, of which trends were also observed in DT and UT cell lines. Taken together, CE-QqQMS-based metabolomics is promising for elucidating energy metabolic changes caused by the molecular target drug and for profiling tissue samples for searching correlations between certain metabolite sets and clinical parameters. References 1. Makinoshima, H., Takita, M., Matsumoto, S., Yagishita, A., Owada, S., Esumi, H. and Tsuchihara, A. (2014) Epidermal Growth Factor Receptor (EGFR) Signaling Regulates Global Metabolic Pathways in EGFR-mutated Lung Adenocarcinoma. J. Biol. Chem. 9, 2370-2378. 2. Kami, K., Fujimori, T., Sato, H., Sato, M., Yamamoto, H., Ohashi, Y., Sugiyama, N., Ishihama, Y., Onozuka, H., Ochiai, A., Esumi, H., Soga, T., Tomita, M. (2013) Metabolomic Profiling of Lung and Prostate Tumor Tissues by Capillary Electrophoresis Time-of-flight Mass Spectrometry. Metabolomics. 9(2): 444-453. 62 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y8. Merging and sharing metabolomics analysis tools with Galaxy: transparent, reproducible, open ’omics. R. L. Davidson1,2, R. J. M. Weber3, P. Li2, M. R. Viant3,* 1 NERC Biomolecular Analysis Facility - Metabolomics Node (NBAF-B), School of Biosciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK; 2GigaScience,BGI-Hong Kong Co. Ltd, 16 Dai Fu Street, Tai Po Industrial Estate, NT, Hong Kong; 3School of Biosciences, University of Birmingham, Birmingham B15 2TT, UK *E-mail: [email protected] (corresponding author) Abstract ’Omics technologies rely heavily on data and computational analyses. Both of these research objects should be citable and openly available if science is to be reproducible. There has been recent policy in e.g. the US and UK that publicly funded research should deposit data in open repositories. The MetaboLights database (1) is now available for metabolomics data. However, there has been no such promotion of software and workflows. Here we bring a full workflow of metabolomics analysis tools into the Galaxy workflow system (2) for the first time. We have implemented the pipeline for 2 widely used non-targeted modalities, direct infusion mass spectrometry (DIMS) using Fourier transform ion cyclotron resonance and liquid chromatography mass spectrometry (LC-MS). These pipelines have been refined within our laboratory over the last several years, and include preparatory processing of the raw data files (SimStitch (3), XCMS (4)), preparation of the X matrix, e.g. normalisation and scaling, automated multivariate statistical analysis and also metabolite identification (MI-Pack (5)). These pipelines represent the core requirements, from start to end, of a “metabolomics analysis” for either data type. With this we hope to initialise the move towards standardised, sharable, transparent workflows in metabolomics while providing a much more intuitive interface for researchers without programming experience and ultimately providing a platform that can integrate this ‘omics with the many others already existing in the Galaxy environment. References 1. Haug, K et al. Nucl. Acids Res. (2013) doi: 10.1093/nar/gks1004. 2. Goecks, J et al. Genome Biol. (2010) 11(8):R86 3. Southam, A.D. et al. Anal. Chem. (2007) 79, pp. 4595–4602 4. Smith, C.A. et al. Anal. Chem. (2006), 78, pp. 779–787 63 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y9. Computational methods to study the effect of perturbations on metabolic phenotypes S Umashankar1, 2, RBH Williams3, S Swarup1, 2, 3,* 1 Metabolites Biology Lab, Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543; 2 NUS Environmental Research Institute (NERI), National University of Singapore, #02-01, T-Lab Building, 5A Engineering Drive 1, Singapore 117411; 3 Singapore Centre for Environmental Life Sciences Engineering (SCELSE),Nanyang Technological University, SBS-01N-27, 60 Nanyang Drive, Singapore 637551; *E-mail: [email protected] (corresponding author) Presenter: Umashankar Shivshankar ([email protected]) Abstract Organisms respond to genetic or environmental perturbations by modulating their cellular metabolism. Changes to these metabolic processes are elicited through the regulation of gene, protein and metabolite levels. Metabolites which form intermediates or end products of these regulatory processes, can be regarded as the ultimate biochemical phenotype of a system. Analysis of metabolomics data in combination with other omics or metadata provide vital insights on how genetic information and environmental factors can influence these processes. Thus, we developed and applied specific data analysis strategies for: • Identifying non-biological sources of variation in untargeted metabolome surveys using multivariate statistical techniques. A filtering procedure based on the singular value decomposition was developed to remove these batch effects while permitting recovery of signal of biological origin. • An untargeted metabolite survey of algae- Chlorella species, to investigate the strain specific signatures of metabolic reprogramming strategies, and associations between physiochemical measures and metabolic pathways. These findings were used to identify metabolic correlates of biotechnology related traits. • To understand the effect of genetic perturbation on metabolic network of Arabidopsis, a data-dependent multi-omics strategy that combines the genomic potential and gene expression outcomes with comprehensive metabolite profiling was developed. This analysis helped discover coordinated regulation of metabolic pathways associated with innate immunity and secondary metabolism. Furthermore, analysis of metabolic network perturbation caused by overexpression of a metabolic enzyme GLDC, revealed novel metabolic reprogramming strategies identifying the gene responsible for non-small cell lung cancer. References 1. 2. Zhang W, Shyh-Chang N, Yang H, Rai A, Umashankar S, Swarup S, Lim E, Lim B. “Glycine Decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis”. , Cell (2012) Rai A, Umashankar S, Sanjay Swarup. “Plant Metabolomics: From Experimental Design to Knowledge Extraction”. Legume Genomics (2013) 64 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y10. Metabolomics Investigation of Green Algae upon Exposure to Pesticide Yan GAO1,*, Qingsong LIN3, Sam Fong Yau LI 1,2* 1 Department of Chemistry, National University of Singapore, 3 Science Drive 2, Singapore, 117543 2 NUS Environmental Research Institute, 5A Engineering Drive 1, T-Lab Building, National University of Singapore, Singapore 117411 3 Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore, 117543 *E-mail: [email protected] (corresponding author) Abstract Algae are an important component of biological monitoring programs for evaluating water quality because of their short lifecycle, rapid reproduction rate and tendency to be most directly affected by physical and chemical environmental factors1. In this study, metabolomics technique is applied to green algae to investigate metabolites changes upon exposure to pesticide. Different concentrations of diazinon (0.01 ppb, 0.1 ppb and 10 ppm) were used to treat green algae Chlorella Vulgaris at different time points(24 h, 48 h, 72 h and 96 h), respectively. LCMSTOF was used to detect metabolite changes in green algae. Clearly separation between control group and treatment groups (0.01 ppb, 0.1 ppb and 10 ppm) can be observed using PCA plot, but the separation among different treatment groups is not good. This may indicate that even lower diazinon concentration (0.01ppb) can cause metabolite changes. Data analysis of different time points and potential biomarkers is still in progress. References 1. Genevieve M. Carr, James P. Neary etc, Water Quality for Ecosystem and Human Health, 2008, United Nations Environment Programme Global Environment Monitoring System/Water Programme. 65 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y11. A Combination of NMR and LC-MS Reveals Metabolic Differences in Toxin and Non-Toxin Producing Microcystis sp. W.L. Zhang 1,2, B.H. Fu 1, S.F.Y. Li 1,2,3 * 1 Department of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543. 2 NUS Environmental Research Institute, National University of Singapore, 5A Engineering Drive 1, Singapore 117411. 3 School of Environment and Energy, Peking University Shenzhen Graduate School, University Town, Xili, Nanshan District, Shenzhen P.R.C. 518055. *E-mail: [email protected] (corresponding author) Abstract Harmful algal blooms predominately formed by Microcystis sp. which produces potent microcystins (MCs) has become an emerging global health issue in freshwater habitats.[1] Extensive research efforts have been carried out to understand the biological role of MCs and environmental conditions that stimulate toxin productions.[2] In this presentation, a combination of nuclear magnetic resonance spectroscopy (NMR) and liquid chromatography mass spectrometry (LC-MS) was employed to reveal the metabolic differences of toxic and non-toxic Microcystis strains. Under similar growth conditions, the cell density of toxic strains is 2-3 folds lower than non-toxic strains, suggesting that the presence of MCs suppress cell growth. The substantial absence (p<0.001) of carbohydrates (sucrose, maltotriose, glucose and glycogen), and amino acids (lysine, aspartate and glutamate) suggest photoinhibition in toxic strains. As MCs are known to produce reactive oxygen species (ROS), and that ROS are one of the main agents responsible for photoinhibition, we reveal toxin-induced photosynthesis impairment. Photosynthesis perturbation would have influence the toxic strains to seek for alternative energy source. Decrease in fatty acids content in toxic strains indicate that Microcystis sp. undergo βoxidation to generate acetyl-coA, which is later used for the biosynthesis of poly(3hydroxybutyrate), a carbon and energy reserve material found in cyanobacteria. The absence of fatty acids with several phospholipid precursors such as sn-glycerol-3-phosphate, phosphocholine and choline also suggest that the decrease in phospholipids would compromise membrane integrity in Microcystis sp., thus result in lower cell density in toxic strains. References 1. Cong LM, Huang BF, Chen Q, Lu BY, Zhang J, Ren YP (2006) Determination of trace amount of microcystins in water samples using liquid chromatography coupled with triple quadrupole mass spectrometry. Anal Chim Acta 569 (1-2):157-168. 2. Neilan BA, Pearson LA, Muenchhoff J, Moffitt MC, Dittmann E (2013) Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ Microbiol 15 (5):12391253. 66 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y12. QuoA expression in Arabidopsis leads to phenolamide channeling to lignins and coordinated effects at key branch points Amit Rai1, Sheela Reuben1, Sanjay Swarup1, 2, 3* 1 Department of Biological Sciences, National University of Singapore, Singapore ([email protected]; [email protected]; [email protected]); 2Singapore Center for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore ([email protected]); 3NUS Environmental Research Institute, National University of Singapore, Singapore ([email protected]), *E-mail: [email protected] (Presenter); [email protected] (corresponding author) Abstract Metabolic perturbations by a gain-of-function approach serves as a tool to alter steady states levels of metabolites and query network properties without effecting native enzyme complexes. In this study, targeted genetics and metabolomics approach was used to understand the network properties and perturbation response of phenylpropanoid metabolism pathways. A novel quercetin oxidoreductase, QuoA, from Pseudomonas putida, utilizes quercetin, a metabolite from phenylpropanoid network released in rhizosphere, and converts it to naringenin1. Since phenylpropanoid biosynthesis in plants involve formation of quercetin from naringenin, we envisaged that QuoA expression in plants will provide us with a genetic tool to “reverse” this biosynthetic step. Therefore, we created transgenic lines expressing bacterial QuoA in Arabidopsis, thus effectively reversing the biosynthesis of quercetin. We selected QuoA transgenic lines for low, medium, and high expression levels of QuoA RNA, which also had corresponding levels of QuoA activity and hypocotyl coloration resulting from increased anthocyanin accumulation. Stems of all three QuoA lines showed increased lignifications and tensile strength in the same order as of QuoA RNA and activity levels2. Comparing gain-offunction transgenic lines, expressing QuoA with loss-of-function line at systems level revealed, (i) metabolite flux from phenolamide into lignin pathways resulted in lignification and increased stem stiffness and (ii) co-ordinated metabolite-to-gene-expression regulation at four key branch points in the phenylpropanoid network. This approach provides dual benefit of imparting stem strength through lignification and improved stress tolerance by up-regulation of flavonoids at the same time. References 1. Pillai BV, Swarup S. 2002. Elucidation of the flavonoid catabolism pathway in Pseudomonas putida PML2 by comparative metabolic profiling. Applied and Environmental Microbiology. 2. Reuben S, Rai A, Pillai BV, Rodrigues A, Swarup S. 2013. A bacterial quercetin oxidoreductase QuoA-mediated perturbation in the phenylpropanoid metabolic network increases lignification with a concomitant decrease in phenolamides in Arabidopsis. Journal of Experimental Botany. 67 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y13. Optimization of metabolite extraction in Pseudomonas aeruginosa PAO1 transition from planktonic to biofilms Saravanan Periasamy1, Victor J. Nesati1, 4, Harikrishnan A.S. Nair1, 3, Staffan Kjelleberg1, 2, 5 , Sanjay Swarup1, 4, and Scott A. Rice1, 2, 5*, 1 Singapore Centre on Environmental Life Sciences Engineering and 2School of Biological Sciences, Interdisciplinary Graduate School, Nanyang Technological University, Singapore, 4 NUS Environmental Research Institute, 5A Engineering Drive 1, T-Lab Building, National University of Singapore, Singapore 117411,5Centre for Marine Bio-Innovation and School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney 2052, Australia. *E-mail: [email protected] (corresponding author) 3 Abstract Bacteria predominantly grow as surface attached biofilms and, just like other surface attached organisms, such as corals, bacteria undergo dispersal events to allow them to seek out and colonize new sites. In this way, bacteria display a biphasic lifecycle, free swimming planktonic cells (dispersal propagules) and surface attached communities. Not surprisingly, bacteria have been shown to express different genes and proteins in the two life phases. Biofilms are important because they impart increased stress tolerance to those microbial populations and communities, including the widely reported increase in antibiotic resistance. As a consequence of their enhanced survival, biofilms pose significant risks where they are responsible for more than 80% of infections and are common problems in a range of industrial settings. Thus, there is a strong motivation to determine how and why bacteria form biofilms and to understand how biofilms are functionally different from planktonic cells. Recent advances in metabolomics methods now allows for detailed studies of the physiology of the populations in both the planktonic and biofilm stages of growth which may provide invaluable information on differences between these two states. Here, we compare the metabolomes of planktonic and biofilm populations of Pseudomonas aeruginosa PAO1. To facilitate these studies, we have further included the wild-type PAO1 and an isogenic wspF mutant that overproduces c-di-GMP and is a hyperbiofilm forming strain. For metabolomics comparison of biofilms and planktonic cells, we first evaluated several extraction methods, including I) Heat Ethanol and Water (HEW), II) Methanol, Chloroform and Water (MCW) (50:50), III) Acetonitrile Methanol and Water (AMW) (40:40:20). Liquid chromatography coupled to mass spectrometry (LC-MS) was used for an untargeted survey of the global metabolites as well as a targeted approach, focused on nucleotides for the comparison of planktonic culture and biofilms. A principal component, multivariate analysis was used to compare samples and to identify features of interest for further interrogation. As expected, the metabolite profiles of the planktonic cells were significantly different from the biofilm populations. For example, both wild type PAO1 and wspF mutant strains showed distinct metabolic profiles between planktonic and biofilm states. Further, results showed that metabolite profiles for biofilms of the wild-type and the wspF mutant indicated the two strains show distinct metabolite patterns. For example, 25 metabolite features were significantly different between the two strains. Metabolites that were significantly higher in the biofilm included 20 metabolite features, suggesting that the purine biosynthetic pathway was highly induced during biofilm growth. Our data demonstrated that distinct metabolites features were play an important role in cellular metabolism during biofilm formation. 68 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 References 1. Gjersing, E., Herberg, J.L., Horn, J., Schaldach, C.M., and Maxwell, R.S. (2007) NMR Metabolomics of planktonic and Biofilm modes of growth in Pseudomonas aeruginosa. Analytical Chemistry 79, 8037-8045. 2. Koex, M.M., Muilwijk, B, Vander werf, M.J., and Hankemeier, T. (2006) Microbial metabolomics with gas chromatography/Mass spectrometry. Analytical Chemistry 78, 1272-1281. 3. Cordell, R.L., Hill, S.J., Ortor, C.A., and Barretr, D.A. (2008) Quatitative profiling of nucleotides and related phosphate-contating metabolites in cultured mammalian cells by liquid chromatography tandem electrospray mass spectrometry. Journal of Chromatography 871, 115-124. 69 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y14. Time-course targeted metabolite profiling in Vitis vinifera cell suspension cultures Nay Min Min Thaw Saw1,*, Claudio Moser2, Pietro Franceschi1 1 Biostatistics and Data Management Group, Computational Biology, Fondazione Edmund Mach, Via Mach1, 38010 San Michele all’ Adige (TN), Italy 2 Gene Functions, Genomics and Biology of Fruit Crop Department, Fondazione Edmund Mach, Via Mach1, 38010 San Michele all’ Adige (TN), Italy *E-mail: [email protected] (corresponding author) Abstract Grapes (Vitis vinifera) are rich sources of phenolic compounds including flavonoids and non-flavonoids. Cell suspension cultures of grapes can be used as a model system for investigating different factors that are able to induce and/or modify secondary metabolite biosynthesis. The characterization of the full time-course metabolic profile of the grape suspension is expected to give by far more information than measurements at one or two timepoints. In this communication we report on the optimization of a complete analytical protocol for the time-course targeted metabolite profiling of Vitis vinifera cv. Pinot Noir suspension cultures. Cell cultures of V. vinifera were grown for 15 days and the sampling was performed daily. The experiment was performed in triplicates. The freeze-dried samples were extracted twice using methanol with the help of sonication and subsequent centrifugation. The identification and quantification of targeted metabolomics in the extract was performed by LC-MS/MS (Vrhovsek et al, 2012) and NMR. Preliminary results indicate that at day seven the most abundant metabolites were benzoic acid derivatives such as 4-aminobenzoic acid, vanillin and vanillic acid, and stilbenes as well as flavan-3-ols and flavonols. Quercetin-type flavonols were found dominantly in all the days during the growth curve. In addition, hesperidin, sinensetin and luteolin were detected as major flavanones in the cell cultures. References 1. Urska Vrhovsek, Domenico Masuero, Mattia Gasperotti, Pietro Franceschi, Lorenzo Caputi, Roberto Viola, Fulvio Mattivi (2012). A Versatile Targeted Metabolomics Method for the Rapid Quantification of Multiple Classes of Phenolics in Fruits and Beverages. Journal of Agricultural and Food Chemistry, 8831-8840. 70 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y15. Metabolomics-based multi-omics analysis of Wolfberry for optimal IBD management Wanping Aw1, 2, Huijuan Jia1, Shinji Fukuda2, Manaka Hanate1, Masaru Tomita2 and Hisanori Kato1 1. Corporate Sponsored Research Program “Food for Life”, Organization for Interdisciplinary Research Projects, The University of Tokyo 2. Institute for Advanced Biosciences, Keio University, Yamagata, Japan Presenting Author Wanping Aw, Ph.D. Email: [email protected] Mailing Address: Keio SFC Systems Biology Laboratory, Kugenumaishigami 1-13-6, Fujisawa, Kanagawa, 251-0022, Japan Abstract Background and Objective Wolfberry (WOL, Lycium barbarum), a traditional Chinese medicinal food, has been reported to have antiaging, anticancer, health-promoting, and immune-boosting properties (1). We aimed to elucidate the anti-inflammatory molecular mechanisms of WOL in a dextran sodium sulphate (DSS) induced colitis model. Methods Seven-week-old male C57BL/6J mice were fed either 2% WOL or control diet for 1 week after which colitis was induced by administering 1.5% (w/v) DSS for 9 days and Disease Activity Index (DAI) was observed. Colonic and hepatic microarray (Mouse Genome 230 2.0, Affymetrix); quantitative iTRAQ hepatic proteome; and capillary electrophoresis-mass spectrometry (CE-MS, Agilent)-based metabolome analysis of plasma and liver metabolites were performed. Results and Discussion WOL significantly suppressed colon length decrease and DAI. Transcriptome and proteome analysis have revealed their common patterns in the suppression of anti-inflammatory cytokines by WOL supplementation. Unique to proteome and metabolome analysis were the amelioration of oxidative markers. These clues from each analysis complement each other, suggesting WOL as an effective nutraceutical in IBD management. Conclusion These findings will serve as a clarion call to promote integrated omics nutrition research to propose dietary-intervention strategies to recover normal homeostasis in disease states and to maintain healthy well-being via daily nutritional consumption. References 1. Tang. WM., et al. A review of the anticancer and immunomodulatory effects of Lycium barbarum fruit. Inflammopharmacology. 20 (6), 307-314 (2012) 2. Amagase. H and Farnsworth NR. A review of botanical characteristics, phytochemistry, clinical relevance in efficacy and safety of Lycium barbarum fruit (Goji). Food Res Int. 44 (7), 1702-1717 (2011) 71 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Y16. Unraveling the metabolic strategies underlying pathogenesis in cereal killer. 1* 1 2 2 Rajesh. N. Patkar , Z. Qu , P. Benke , S. Swarup , N. Naqvi1 1 Temasek Life Sciences Laboratory (TLL), 1 Research Link, Singapore 2 NUS Environmental Research Institute (NERI), Singapore *E-‐mail: [email protected] (presenting author) Abstract Blast is a fungal disease of a wide variety of food crops including rice, wheat and barley. Significant yield losses due to Blast disease make the causal fungal pathogen Magnaporthe oryzae a serious threat to global food security. M. oryzae has a remarkable ability to modulate the host immunity for its own advantage. Here, we found that loss of an Antibiotic Biosynthesis Monooxygenase (Abm), from a secondary metabolism gene cluster, led to fungal inability to suppress host innate immunity, and resulted in non-‐pathogenicity in the mutant. Using a plant-‐based assay combined with chemical analysis tools, we identified that Abm directly inactivates jasmonic acid (JA) into 12-‐hydroxyjasmonic acid (12-‐OH-‐JA) to down-‐regulate the defense response in the host. Further systematic characterization showed that M. oryzae expresses Abm intracellularly to synthesize endogenous 12-‐OH-‐JA, which is secreted out at the time of invasion, most likely to suppress early wounding response in the host plant. Interestingly, M. oryzae secreted Abm upon invasion, likely to suppress the plant innate immunity by converting host JA to 12-‐OH-‐JA therein. This study not only suggests fungal Abm as a potential fungicidal target towards disease control and food security, but also provides a synthetic strategy for transformation of a versatile small-‐ molecule plant hormone. 72 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Agilent Sponsored Talk Two-dimensional liquid chromatography: a new technique for metabolomics? O.A.H.Jones1*, 1 School of Applied Sciences, RMIT University, GPO Box 2476, Melbourne, VIC 3001, Australia * E-mail: [email protected] Abstract Multi-dimensional chromatography is a system in which more than one mechanism of separation is applied to a sample, with each mechanism being considered an independent separation dimension (1). Multi-dimensional gas chromatography is an established technique with a distinguished history in metabolomics. However, as with all forms of GC, samples are limited to those compounds that are, or can be made, volatile. In contrast, two-dimensional liquid chromatography (LC×LC) is a relatively new technique and does not require derivatisation. We have applied this method to the analysis of complex samples and demonstrate that the enhanced resolution and peak capacity allow peaks to be detected that could not be separated using standard, one-dimensional LC. An offline reversed phase x reversed phase LCxLC method was developed and applied to Agaricus bisporus mushrooms in order to demonstrate the potential of the technique and assess the effect of UV irradiation on the mushroom's metabolic profile. The method allowed the detection of 158 peaks in a single analytical run and aa total of 51 compounds including sugars, amino acids, organic and fatty acids and phenolic compounds were identified. After irradiation of the mushrooms with UV for 30 s the number of peaks detected decreased from 158 to 150; 47 compounds increased in concentration while 72 substances decreased. This is the first time that two-dimensional liquid chromatography has been carried out for the metabolomic analysis of mushrooms. The data provide an overview of the gain/loss of nutritional value of mushrooms following UV exposure and demonstrate that the increased peak capacity and separation space of LCxLC has great potential in metabolomics. References 1. Stevenson, P.G., M. Mnatsakanyan, G. Guiochon, and R.A. Shalliker, (2010) Peak Picking And The Assessment Of Separation Performance In Two-Dimensional High Performance Liquid Chromatography. Analyst 135, p. 1541-1550. 73 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Technical Site Visit Information Agilent LC/MS facility Agilent Technologies Inc. (NYSE: A) is the world's premier measurement company and a technology leader in chemical analysis, life sciences, electronics and communications. The company's 18,500 employees serve customers in more than 100 countries. The company is a trusted partner to thousands of the scientists and lab analysts worldwide who are focused on basic research, drug discovery & development, food & environmental testing, forensics and clinical research. The Agilent LC/MS facility is located within the Agilent Life Sciences Group Order Fulfillment Center, located at its Yishun, Singapore, site. The center will be responsible for efficient management of the supply-chain network -- including planning, procurement, value-added engineering and logistics. The center ensures that the LC/MS facility is able to efficiently and effectively deliver configurable solutions to customers worldwide while maintaining Agilent's hallmark of quality. Information about Agilent is available at www.agilent.com. 74 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 NERI Laboratory at T-Lab and CREATE. Laboratory and Technical Staff Support The NERI Laboratory serves as a strong backbone analytical support for NUS researchers in environmental research. It collaborates closely with different initiatives and programmes as well as leading analytical equipment industries to bring the latest advanced analytical and spectroscopic instruments. The instruments will allow researchers to unravel and discover mysteries in their respective research and identify traces of organic and inorganic compounds that may be previously undetected. Also, the NERI Laboratory has a comprehensive sample preparation facility that expedites the clean-up and extraction from complex sample matrices. The NERI Laboratory is managed by a team of dedicated and experienced technical and research staff. They provide expertise in assisting the NUS community and industry engaged with NERI doing environmental research. They are also committed in ensuring a high standard of occupational safety and health for staff and researchers. Exchange of Technical Knowledge As a focal point for NUS researchers on environmental research, NERI's laboratory is also a facility where researchers from multi-disciplines (biologists, bioinformatics engineers, analytical chemists, toxicologists, chemical and biomolecular engineers and environmental engineers) gather and exchange technical knowledge to enhance research in their respective fields. The more experienced research staff will often interact with postgraduates to provide guidance and advice in their respective research studies. Information about NERI is available at: http://www.nus.edu.sg/neri/index.html 75 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Sponsors’ Profile The Merlion Metabolomics Workshop Organising Committee gratefully acknowledges the generous support from the following sponsors: 76 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Agilent Technologies Inc. (NYSE: A) is a leader in chemical analysis, life sciences, diagnostics, electronics and communications. The company’s 20,600 employees serve customers in more than 100 countries. Agilent had revenues of $6.8 billion in fiscal 2013. Information about Agilent is available at www.agilent.com. Singapore is the hub for Agilent’s infrastructure, strategic supply chain, global logistics, treasury, business support as well as sales and marketing services functions. The site also houses the life sciences manufacturing facility. The facility was officially opened in 2010 by Minister, Prime Minister’s Office, Mr. S. Iswaran, and Agilent CEO, Mr. Bill Sullivan. The manufacturing facility is ISO 9001 certified, ISO 13485 certified in FDA Class 1, in addition to certifications from Korean FDA and Singapore HSA, it is also QSR compliant. The manufacturing facility produces Liquid Chromatography Mass Spectrometers (LC/MS), Automation, Supercritical Fluid Chromatography (SFC) and Microarray Scanners, which are shipped to customers globally. 77 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Waters Corporation is a publicly traded laboratory analytical instrument and software company headquartered in Milford, Massachusetts. The company employs more than 5,000 people, with manufacturing facilities located in Milford, Taunton, Massachusetts; Wexford, Ireland; Manchester, England; and contract manufacturing in Singapore. Waters markets to the laboratory-dependent organization in these market areas: liquid chromatography, mass spectrometry, supercritical fluid chromatography, laboratory informatics, rheometry and microcalorimetry. Website: http://www.waters.com/waters/home.htm?locale=en_US Under the Thermo Scientific brand of Thermo Fisher Scientific, Inc. (NYSE:TMO), we help scientists meet the challenges they face in the lab or in the field every day. From routine analysis to new discoveries, our innovations help professionals do the science they need to do, the way they want to do it. Our high-end analytical instruments, laboratory equipment, software, services, consumables and reagents help our customers solve complex analytical challenges, improve patient diagnostics and increase laboratory productivity. Website: http://www.thermoscientific.com/en/home.html 78 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Location Map of NUS-University Town Visitors can get to University Town (UTown) by: A. MRT & Bus From Buona Vista MRT Station - Bus 196 Walk to the bus stop opposite Buona Vista MRT Station located near Exit D, Bus Stop No. 11369. Board Bus 196 (towards Clementi Interchange). Alight 6 stops later at Bus Stop No. 19059. Walk approximately 5 to 8 minutes to UTown. From Clementi MRT Station - Bus 183 Walk to Clementi Bus Stop, Bus Stop No. 17179, from Clementi MRT Station (Exit B). Board Bus 183 (towards Jurong East Temp Interchange). Alight 3 stops later at Bus Stop No. 17099. Walk approximately 8 to 10 minutes to UTown. From Clementi Bus Interchange - Bus 96 Board Bus 96 (towards Clementi Interchange). Alight 3 stops later at Bus Stop No. 17099. Walk approximately 8 to 10 minutes to UTown. From Kent Ridge MRT Station - NUS Internal Bus Shuttle Walk to Kent Ridge Bus Stop, Bus Stop No. 18331, from Kent Ridge MRT Station (Exit A). Board NUS Internal Bus Shuttle D2. Alight 5 stops later at UTown. B. NUS Internal Bus Shuttle Services D1 and D2 79 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 Internal Shuttle Bus D1/D2 80 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 NOTES _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ 81 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ 82 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ 83 MERLION METABOLOMICS WORKSHOP SINGAPORE 2014 _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ _________________________________ 84 Supported by: Silver Sponsor: Sponsors:
© Copyright 2026