SULSA Drug Discovery Capacity Dr Stuart McElroy Dundee Drug Discovery Unit
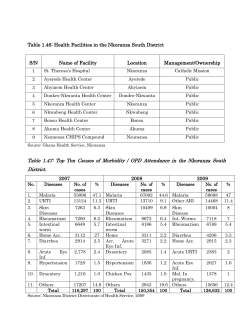
SULSA Drug Discovery Capacity Dr Stuart McElroy Dundee Drug Discovery Unit What is SULSA? Translational Biology Systems Biology Cell Biology High Throughput Screening (HTS) and High Throughput Screening (HTS) and Chemistry Chemistry Catalyst funds Catalyst funds • SULSA has focused a proportion of funds to pump-prime HTS and Chemistry Catalysts projects (£20K per project) • Projects come from SULSA Universities • Projects are bid for competitively • Projects are associated with SULSA drug discovery facilities • Process: Hit – Lead – Validate • Aim: De-risk HTS hits to make them more attractive for further investment • Out-comes so far: • Two major licensing deals with a major pharmaceutical company • Several follow-on grants from charities and research councils SULSA Shared Facilities SULSA Shared Facilities Sequencing GenePool Biomedical Imaging Metabolomics OMX Microscope Electron Cryomicroscopy SHWFGC Proteomics ScotMET HTP BioScreening SSPF ARKGenomics Bioinformatics Drug Screening Scottish Hit Discovery Facility Drug Discovery Portal Scottish Bioscreening Facility Bioworkstation Scottish Biologics Facility Positron Emission Tomography Marine Biodiscovery Centre IVIS Spectrum Imager CTCB SIDR Other Micro and Nano Fabrication Transgenics Facility Data analysis Group (DAG) DDU Screening Facilities • Drug Discovery Portal – – – • Strathclyde Natural Products Library – – – • – – – – – High Content Image Based Screening (HCS) with the GE IN Cell 2000©. High throughput image capture and analysis of numerous parameters in a biologically relevant context (e.g. whole cells). Automated microscopy (uni-well, 96, 384, 1536 well) Marine Biodiscovery Centre . – – – • Biologic ligands (classical monoclonal antibodies and their fragments) Peptides and other binding scaffolds as tools for biomarker validation diagnostic assay development, in vivo imaging and drug discovery.. Scottish Bio-screening Facility (SBF) – • Natural Products Library of 5120 extracts from around the world Covers 90% of plant families - One of the most bio-diverse (and hence chemically diverse) collections available for screening. access to the Maybridge Hitfinder ™ compound collection of 14,000 pure compounds specially selected for their chemical diversity and drug-like properties. Scottish Biologics Facility – • Collection of diverse compounds that have been synthesised in-house In silico screening that is linked to the originating chemists for synthesis Physical screening and study of structure-activity relationships High Content Image Based Screening (HCS) with the GE IN Cell 2000©. High throughput image capture and analysis of numerous parameters in a biologically relevant context (e.g. whole cells). Automated microscopy (uni-well, 96, 384, 1536 well) Drug Discovery Unit (DDU) Drug Discovery Facilities Scottish Biologics Facility www.abdn.ac.uk/sbf Marine Biodiscovery Centre www.abdn.ac.uk/ncs/chemistry/res earch/mbc Scottish Bio-screening Facility (SBF) www.gla.ac.uk/researchinstitutes/i ii/facilities/imaging/sbfglasgow Drug Discovery Portal www.ddp.strath.ac.uk Strathclyde Natural Products Library Drug Discovery Unit (DDU) http://www.drugdiscovery. dundee.ac.uk/ Early Stage Drug Discovery engine ● ● ● ~40 staff with >170 years collective BioPharma drug discovery experience DDU Technologies • In house designed and selected screening sets – – – – – Diversity sets >63,000 and >16,000 compounds Focused sets, incl. kinase, ion channel, epigenetic, nuclear hormone receptor Fragment set Bioactive compounds - Prestwick Chemical Library, Biomol, Tocris Human genome-wide RNAi library Inert environment compound storage • Industry standard:• – Data management – ActivityBase, Dotmatics Browser – Liquid handling robotics and breadth of assay formats • including surface plasmon resonance and high content screening – Assay data quality control – Computational chemistry tools – Parallel synthetic chemistry capabilities • DMPK capabilities – In vitro metabolic stability, CYP450 inhibition, plasma protein and brain tissue binding, in vivo DMPK, in vivo Pgp substrate assessment. – Waters QUATTRO PREMIER XE LCMSMS • PK/PD efficacy models. Ways of working • Collaboration • Fee for service – IP is wholly owned by commissioning organisation • Partnership – Matching funds – Shared risk, shared rewards Hits to Leads - Premature stop codons >1,800 diseases where a PTC is the cause Monogenic disease Polygenic disease Cystic fibrosis 1 in 34,000 Atopic disease Hurlers Syndrome 1 in 100,000 Diabetes Cancer Adrenoleukodystrophy 1 in 20,000 Recessive Dystrophic Epidermolysis Bullosa (RDEB) • Mutation within COL7A1, which connects epidermis with dermis. • Minor mechanical forces upon the skin causes severe blistering, ulcers and scarring, which leads to fusion of fingers and toes, SCC etc… • Mouth and esophagus are also affected. (difficult to chew and swallow foods) Maisy – RDEB patient Produced with permission from DebRAUK Screening Data Read-Through Assay Primary Screen Frequency Histogram f.Luc 3000 5’ CAG GCA TGA GAC AGC 3’ Q A X No of Compounds 2500 2000 1500 1000 500 0 120 440 760 1080 1400 1720 Signal Range (RLUs) 2040 2360 2680 3000 Hit Validation GCA-TGA-GAC A X D K6a cDNA YFP CMV promoter K6aYFP wild type K6aYFP geneticin (100µg/ml) 1:50 1:2 negative control (1% DMSO) Hit Validation GCA-TGA-GAC A X D K6a cDNA YFP CMV promoter K6aYFP wild type K6aYFP geneticin (100µg/ml) 1:50 1:2 negative control (1% DMSO) Stop codon read through profile * (30uM) TGA G C T A TAG G C T A TAA G C T A Geneticin DMSO (200uM) Read through of natural stops? Natural stop codon (TAAA) Geneticin 20nM 200nM 2uM 20uM (100ug/ml) No peptide downstream of the natural stop codon was detected. Natural stop codon (TAAA) Patient-derived Cell lines Current status • Partnered with GlaxoSmithKline Malaria • Caused by 5 species of Plasmodium. – P. falciparum most pathogenic. – P. vivax leads to latent tissue forms and recurrence. • >250 million clinical cases per year. • ~1 million deaths/ annum (mainly children in subSaharan Africa). • Estimated cost to African economy $12 billion/ annum. • Urgent need for new drugs: – Tackle resistance of existing drugs. – Develop new indications. Animated lifecycle of the malaria parasite TRANSMISSION TO MAN TRANSMISSION TO MAN LIVER Sporozoites Nucleus Hypnozoite Sporozoites 15-30 mins P. vivax dormant stage Oocysts 9-12 days Infected Hepatocyte Ookinete Diploid Zygote 12-36h Macrogametocyte (Exflagellation) 1h 5.4 days Schizont Macrogametocyte 15 mins Merozoites 9 days Erythrocyte TRANSMISSION TO MOSQUITO Gametocytes Ring 43 – 48 h Cycle leading to clinical symptoms Schizont 1 Trophozoite What does a new anti-malarial drug need to look like? • • • • • Cost < $1 for a complete treatment. Very safe - suitable for children and pregnant women. Available in tablet form. Stable in tropical conditions. Various Different Requirements (Target Product Profiles) 1. Single exposure radical cure for uncomplicated malaria 2. Non-artemesinin-based combination therapy for uncomplicated malaria 3. Non-oral treatment for cerebral malaria. 4. Single agent for radical cure of vivax malaria. 5. Single agent which can prevent transmission of malaria. 6. Intermittent preventative treatment of malaria in pregnant women. 7. Chemoprophylaxis of malaria. Our starting point • Carried out phenotypic screen of our in-house kinase library. • Screened 4731 compounds against P. falciparum at 3 µM. • Rescreening of the hits gave 21 compounds with EC50 < 1 µM. • Identified a number of series – focused effort on 3 series. Progress with MMV04 series. EARLY LEADS CRITERIA EC50 vs. Pf(3D7) April 2010 <0.1 0.12 > 10 fold 20 (µM) EC50 vs. MRC5 (µM) logP <5 logD pH 7.4 <5 4.3 Mouse Cli (ml/min/g) Human Cli (mL/min/g) Mouse PPB % Stable 5.3 Solubility (µM) >10µM In vivo efficacy Stable <99% Depression of parasitaemia orally 99.7 Progress with MMV04 series. EARLY LEADS April 2010 Oct 2010 <0.1 0.12 0.27 > 10 fold 20 >50 CRITERIA EC50 vs. Pf(3D7) (µM) EC50 vs. MRC5 (µM) logP <5 logD pH 7.4 <5 4.3 3.9 Mouse Cli (ml/min/g) Human Cli (mL/min/g) Mouse PPB % Stable 5.3 1.5 99.7 97.2 Solubility (µM) >10µM In vivo efficacy 3.9 Stable <99% Depression of parasitaemia orally 13 Progress with MMV04 series. EARLY LEADS April 2010 Oct 2010 April 2011 <0.1 0.12 0.27 0.05 > 10 fold 20 >50 47 3.9 2.1 CRITERIA EC50 vs. Pf(3D7) (µM) EC50 vs. MRC5 (µM) logP <5 logD pH 7.4 <5 4.3 3.9 1.7 Mouse Cli (ml/min/g) Human Cli (mL/min/g) Mouse PPB % Stable 5.3 1.5 0.8 Solubility (µM) >10µM In vivo efficacy <0.5 Stable <99% Depression of parasitaemia orally 99.7 97.2 59 13 >230 Yes; 99% reduction in parasitaemia; 25mg/kg bid PO In vivo DMPK Profile • Good oral bioavailability and long half life in mouse and rat. • In vivo PK consistent with once daily oral dosing Mouse Rat Half life 19 h 18 h Oral Bioavailability 74% 84% EC99 EC99 Humanised mouse model (P. falciparum) DDD107498 efficacy in Plasmodium falciparum model Immunocompromised mice with human erythrocytes, capable of being infected with P. falciparum DDD00107498 (mg/Kg) Treatment 10 0 % parasitemia 0.05 1 ED90 0.1 0.3 0.6 0.1 1 3 Limit of detection 0.01 0 1 2 3 4 5 6 7 Day after infection ED90 0.6-1 mg/Kg Summary of lead compound • Good Efficacy – ED90 0.1-0.3 mg/kg 4 x qd po (P. berghei model) – Curative at 10 mg/kg 4 x qd po (P. berghei model) – ED90 0.6 mg/kg 1 x qd po. Partial cures at 30 mg/kg (P. berghei model) – ED90 0.6-1 mg/kg 4 x qd po (SCID Pf model) • Good pharmacokinetics – Long T½ (mouse/rat) – High bioavailability (70-80% mouse/rat) – Predicted human dose: 24-45 mg/day and half-life ~ 40h • No toxicological concerns – – – – No mutagenicity in Ames No hERG liability No concerns from receptor panel No adverse events observed in mice at 100 x ED90 • Proposed TPP – This is still being investigated – Could include single dose treatment Current Status • Late Lead declared • Candidate selection scheduled for July Summary • SULSA is a research pooling partnership between the major Scottish universities aimed at maintaining and advancing Scotland’s global position in the Life Sciences by recruiting international research leaders and funding world-class research facilities • The DDU has been able to advance difficult to fund science through funding mechanisms such as SULSA • Investment in translational early stage drug discovery/chemical biology is essential to progress risky but under exploited basic research and neglected therapeutic areas Acknowledgements • Read Through Project Team: Irwin McLean Toshifumi Nomura Ulrike Gartner Emma Warbrick Leah Torrie Andrew Woodland Gavin Wood Julie Frearson • DDU MMV Project Team: Beatriz Baragaña Neil Norcross Irene Hallyburton Suzanne Norval John Thomas Achim Porzelle Caroline Wilkinson • DDU Management Team: Ian Gilbert Kevin Read David Gray Alan Fairlamb Paul Wyatt • DMPK: Robert Kime Laste Stojanovski Fred Simeons • Ngai Mok for computational chemistry • Daniel James and Alistair Pate for data management • Medicines for Malaria Venture • Dr Paul Willis • Dr Simon Campbell
© Copyright 2026