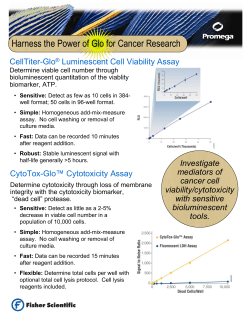
User Guide iPLEX Pro Sample ID Panel ®
User Guide iPLEX® Pro Sample ID Panel The iPLEX Pro Sample ID Panel is For Research Use Only. Not For Use in Diagnostic Procedures. iPLEX ® Pro Sample ID Panel User Guide DOC. SQNM-USG-CUS-050 CO 12-579 Version 1.0 11 January 2013 TRADEMARKS MassARRAY, MassEXTEND, iPLEX, SensiPLEX, SEQUENOM, and SpectroCHIP are registered trademarks of Sequenom, Inc. EpiTYPER, MassCLEAVE, SEQureDX, TypePLEX and any other marks so indicated, are trademarks of Sequenom, Inc. All other trademarks or service marks set forth herein are the property of their respective owners. PATENTS Sequenom's patented nucleic acid analysis by mass spectrometry methods and products are protected under United States patent rights including but not limited to 5,869,242; 6,024,925; 6,238,871; 6,258,538; 6,300,076; 6,440,705;6,500,621; 6,558,623; 6,569,385; 6,979,425; 6,994,969; 7,025,933; 7,285,422; 7,332,275; 7,390,672; 7,419,787; and 7,501,251 and patents pending including but not limited to 20040081993A1, 11/089,805, and all of the foreign equivalent patent rights of the foregoing. COPYRIGHT © 2012. All rights reserved. No part of this publication may be reproduced, distributed, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, or stored in a database or retrieval system, for any reason other than a licensee's internal use without the prior written permission of SEQUENOM. Printed in the United States of America. i Chapter 1 Introduction Introduction . . . . . . . . . . . . . . . . . . . . . . . .1 . . . . . . . . . . . . . . . . . . . . . . .1 Software Installation . . . . . . . . . . . . . . . . . . . . .2 Contacting SEQUENOM . . . . . . . . . . . . . . . . . . . .2 Chapter 2 Workflow Overview and Inventory Checklist . . . . . . . . . . 5 Workflow Overview . . . . . . . . . . . . . . . . . . . . .5 Inventory Checklist . . . . . . . . . . . . . . . . . . . . . .5 Laboratory Work Areas . . . . . . . . . . . . . . . . . . . .7 Chapter 3 Configuring the Software . . . . . . . . . . . . . . . . . . 9 Importing the Sample ID Assay Group File . . . . . . . . . . . . . Setting Up Sample Identification . . . . . . . . . . . . 10 . . . . .9 Chapter 4 Assay Protocol . . . . . . . . . . . . . . . . . . . . . 17 Isolating DNA from Samples . . . . . . . . . . . . . . . . . 17 Performing PCR Amplification . . . . . . . . . . . . . . . . . 17 Performing the SAP Treatment . . . . . . . . . . . . . . . . . 19 Performing the iPLEX Pro Extend Reaction . . . . . . . . . . . . . 20 Desalting the Extend Reaction . . . . . . . . . . . . . 22 Designing a Sample ID Plate in PlateEditor . . . . . . . . . . . . . 23 . . . . Chapter 5 Dispensing Samples to a SpectroCHIP® Array . . . . . . . . . . . . . . . . . . . 25 Preparing the Nanodispenser and Reaction Plate Selecting Nanodispenser Settings . . . . Loading the Reaction Plate and SpectroCHIP . . . . . . . . . . . 25 . . . . . . . . . . . . 26 . . . . . . . . . . . . 26 Starting the Dispensing Run . . . . . . . . . . . . . . . . . . 26 Removing the SpectroCHIP . . . . . . . . . . . . . . . . . . 27 Chapter 6 Acquiring Data . . . . . . . . . . . . . . . . . . . . . 29 Starting the MassARRAY Analyzer Software . . . . . . . . . . . . 29 Loading the SpectroCHIP Array(s) . . . . . . . . . . . . . 29 Creating an Input File using Typer Chip Linker . . . . . . . . . . . . 30 Setting Up the Automatic Run and Checking the Barcode Report . . . . . . 31 Starting the Automatic Run . . . . . . . . . . . . . . . . . . . 32 Unloading the SpectroCHIP Array(s) . . . . . . . . . . . . . . . 33 iPLEX® Pro Sample ID Panel User Guide . . SQNM-USG-CUS-050 ii Chapter 7 Analyzing Data . . . . . . . . . . . . . . . . . . . . . 35 Generating Sample ID Reports . . . . . . . . . . . . . . . . . 35 Reading Sample ID Reports . . . . . . . . . . . . . . . . . 39 . Appendix A Assay Information . . . . . . . . . . . . . . . . . . . . 49 Appendix B UNG PCR Protocol Appendix C Troubleshooting Appendix D Licensing . . . . . . . . . . . . . . . . . . . 51 . . . . . . . . . . . . . . . . . . . . 53 . . . . . . . . . . . . . . . . . . . . . . . 55 iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 1 Introduction Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1 Software Installation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 Contacting SEQUENOM . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 1.1 Introduction The iPLEX® Pro Sample ID Panel is designed to screen DNA extracted from FFPE samples, cell lines, and/or tissues in order to identify sample mismatch, sample duplication, and/or sample identification, and to quantify input DNA prior to further genotyping assays. It is intended for research use only and is not for use in diagnostic procedures. The panel uses Sequenom’s iPLEX Pro biochemistry with a specific Sample ID oligo mix. After spectra are obtained on the MassARRAY ® Analyzer, the Typer Sample ID software performs sample quality control, determines the gender of the sample based on three gender assays, does a pairwise comparison of all samples across a set of 44 SNPs for sample identification, and generates a results report. (See Appendix A for a list of the assays used.) Sample Quality Control The Sample ID software performs a quality control check on each sample by: • Calculating the number of amplifiable copies of DNA, based on five quantitative assays. Samples with fewer than 500 amplifiable copies will fail quality control. • Counting the number of successful SNP calls made. Samples with fewer than 30 successful SNP calls will fail quality control. Sample Matching After quality control, the Sample ID software performs a pairwise comparison of all valid samples across a set of 44 SNPs and determines which samples match each other (i.e., originate from the same person). These comparisons are reported in three categories: • Unexpected mismatch (i.e., two samples were supposed to match but did not). • Unexpected match (i.e., two samples were not supposed to match but did). • Expected match (i.e., two samples were supposed to match and did). Results for non-tumor vs. non-tumor comparisons and non-tumor vs. tumor comparisons will be reported separately in each of these three categories. The vast majority of results are expected mismatches and these results are not explicitly reported by the software. When two samples are compared, each of the 44 pairs of SNP calls are compared individually. If either sample generated a no call, that SNP is ignored for that comparison. If a non-tumor and a tumor sample are being compared, only SNPs that are homozygous for the non-tumor sample are considered, to account for potential loss of heterozygosity in the tumor. If the two SNP calls do not match, an assay-specific penalty is applied to the comparison score between those two samples, with homozygote to homozygote mismatches penalized more heavily than homozygote to heterozygote mismatches iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 2 Chapter 1 Introduction Sample ID Database The Sample ID software allows you to develop a repository of known samples against which future samples can be compared. As samples are run on the MassARRAY system and analyzed on the Typer software, you can choose to add them to the Sample ID database. Then as new samples are run on the system, you can compare them to all previously run samples in your database by choosing to generate a historical report. See page 35 for more information. 1.2 Software Installation The iPLEX Pro Sample ID Panel software requires Typer version 4.0.52 or higher. For instructions on installing Typer 4.0.52, and for information on Oracle client compatibility, see MassARRAY Typer 4.0.52 Release Notes. iPLEX Pro Sample ID Panel reports are generated in both .csv and HTML versions; Internet Explorer or Chrome are required to open the HTML reports. 1.3 Contacting SEQUENOM Please contact your local Sequenom office for customer support. CORPORATE HEADQUARTERS & NORTH AMERICA OFFICE 3595 John Hopkins Court San Diego, CA 92121-1331 USA Phone: 1-858-202-9000 Fax: 1-858-202-9001 Order Desk: 1-858-202-9301 Order Desk Fax: 1-858-202-9220 E-mail: [email protected] Help Desk: 1-877-4GENOME (1-877-443-6663) Help Desk: 1-858-202-9300 E-mail: [email protected] E-mail: [email protected] E-mail: [email protected] EUROPEAN OFFICE Mendelssohnstrasse 15D D-22761, Hamburg Germany Phone: (+49) 40-899676-0 Fax: (+49) 40-899676-10 Order Desk: [email protected] E-mail: [email protected] ASIA PACIFIC OFFICE 300 Herston Road Herston, QLD 4006 Australia Phone: (+61) 3088 1600 Fax: (+61) 3088 1614 E-mail: [email protected] iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Contacting SEQUENOM SEQUENOM, K.K PMO Nihonbashi Odemmacho, Building 5F 6-8 Nihonbashi Odemmacho, Chuo-ku Tokyo 103-0011 Japan Phone: (+81) 3 6231-0727 Fax: (+81) 3 3668-6088 E-mail: [email protected] BEIJING REPRESENTATIVE OFFICE Technology Building, Suite 702B, No. 28, Tian-Zhu Road Tian-Zhu Airport Industrial Zone A Shunyi District Beijing, 101312 China Phone: (+86) 10-8048 0737 Fax: (+86) 10-8048 0740 E-mail: [email protected] iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 3 4 Chapter 1 Introduction iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 2 Workflow Overview and Inventory Checklist Workflow Overview . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5 Inventory Checklist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5 Laboratory Work Areas . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7 2.1 Workflow Overview Running the iPLEX Pro Sample ID Panel involves performing the iPLEX Pro biochemistry, designing a plate in Typer PlateEditor, dispensing samples to a SpectroCHIP, acquiring data on the MassARRAY Analyzer, and running the Sample ID reports in TyperAnalyzer, as outlined in Table 2.1. Table 2.1 Workflow Steps Step Description 1 Import the Sample ID Assay Group file to the Typer software. NOTE: This only needs to be done once, before first use of the Sample ID software. See Page 9 10 2 Set up sample identification conventions in the Sample ID software. NOTE: Only required before first use of the Sample ID software if you want to change the default settings. You need to repeat this step for future runs only if you want to change your naming system. 3 Purify sample DNA from FFPE or fresh tissue or cell line of choice and prepare DNA working dilutions. 17 4 Amplify DNA using the provided iPLEX Pro Sample ID Panel primers. 17 5 Dephosphorylate any remaining free deoxynucleotides in the amplification reaction mixture (SAP treatment). 19 6 Process the iPLEX Pro Sample ID Panel Extend reactions using the provided Extend Primers. 20 7 Desalt the extension products. 22 8 Design a Sample ID plate in Typer PlateEditor. 23 9 Dispense the Extend reaction products to a SpectroCHIP array (chip). 25 10 Acquire data by analyzing the chip on the MassARRAY Analyzer instrument. 29 11 Generate an iPLEX Pro Sample ID Panel assay report in TyperAnalyzer. 35 2.2 Inventory Checklist Kit Contents The following items are included in the iPLEX Pro Sample ID Panel Assay Kit (catalog #25093 for 10 x 96 kit, sufficient for 960 samples; #25094 for 2 x 384 kit, sufficient for 768 samples). Upon receipt, store the items as described. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 6 Chapter 2 Workflow Overview and Inventory Checklist Table 2.2 iPLEX Pro Sample ID Panel Assay Kit Component Storage Conditions Materials Provided Shipping Condition Storage Temperature Quantity iPLEX Pro Sample ID Panel PCR Primer Mix Dry Ice -10 to -25°C 1 iPLEX Pro Sample ID Panel Extend Primer Mix Dry Ice -10 to -25°C 1 iPLEX Pro Sample ID Q-Mix Dry Ice -10 to -25°C 1 Note: Do not subject the Sample ID Q-Mix to more than 5 freeze/ thaw cycles. Sequenom PCR Reagent Set, including: Dry Ice -10 to -25°C 1 iPLEX Pro Reagent Set: Dry Ice iPLEX Pro Reagent Set: -10 to -25 °C SpectroCHIPs and Resin: Room Temperature SpectroCHIPs and Resin: Room Temperature • PCR Accessory Reagents • PCR Enzyme iPLEX Pro Reagent Set, including: • Thermosequenase Enzyme and iPLEX Pro Reagents • SpectroCHIP 384 element arrays (2 arrays, sufficient for 768 samples; cat. # 25094) 1 OR OR 1 • SpectroCHIP 96 element arrays (10 arrays, sufficient for 960 samples; cat. # 25093) • CLEAN Resin Optional: Users wishing to use UNG enzymes for PCR must order the following separately: • Uracil-N-Glycosylase (UNG), 5 U/μl, 2,500 units: cat. no. 1744. • dNTPs, 25 mM: cat. no. 1745. Required Instruments and Equipment Table 2.3 Required Instruments and Equipment Instruments and Equipment Supplier Quantity MassARRAY Analyzer 4 OR MassARRAY Analyzer Compact Sequenom 1 MassARRAY Database Server 1 MassARRAY Nanodispenser Sequenom 1 384-well semi-skirted resin plate Sequenom 1 96-well plate centrifuge Any 1 Microtube centrifuge Any 1 Spectrophotometer for quantifying DNA NanoDrop 1 Standard thermocycler Any 1 OR 96-well semi-skirted resin plate iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Laboratory Work Areas Required Software Table 2.4 Required Software Required Consumables Software Supplier Quantity Typer 4.0.52 or higher (purchased separately) Sequenom 1 ’R’ Environment R–2.9.1 http://cran.r-project.org 1 Table 2.5 Required Consumables Item Supplier Full-skirted 384-well reaction plate Sarstedt or Abgene Quantity 1 Clear adhesive sealing film-PCR CS/100 (for PCR cycling) Myriad Industries part no. 3150-0558 1 Sealing film, non-sterile Myriad Industries part no. 0425-0197 1 Sealing roller tool MJ Research (Bio-Rad) part no. MSR0001 1 Single and multi-channel pipettors (0.5, 20, 200, and 1000 μl) Any 2 sets (pre- and post-PCR) Repeater pipettors Eppendorf Repeater Plus with Combitips Plus 0.1 ml or 0.5 ml 2 sets (pre- and post-PCR) Filtered pipette tips (10, 20, 300, 1000 μl) Any Varies Microtubes (0.5, 1.5, and 2 ml) Any 50 Reagent reservoirs Any 2 Water, HPLC grade J.T. Baker part no. 4218-02 DNA AWAY Molecular Bio Products (MBP) 250 ml 100% ethyl alcohol, HPLC grade EMD part no. EX0276-3 1 gallon OR Un-skirted 96-well reaction plate 1L 2.3 Laboratory Work Areas The laboratory space should include three separate (non-contiguous) work areas to prevent contamination of PCR products. Table 2.6 shows the activities that are conducted in each area. Table 2.6 Lab Area Activities Lab Area Activities 1 Isolation and dilution of DNA. 2 Pre-PCR preparation and addition of DNA template to the PCR cocktail. 3 Post-PCR processing. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 7 8 Chapter 2 Workflow Overview and Inventory Checklist iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 3 Configuring the Software Importing the Sample ID Assay Group File . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9 Setting Up Sample Identification . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10 Before using the Typer software to analyze iPLEX Pro Sample ID Panel data for the first time, you must: • Import the Sample ID assay group file into Typer AssayEditor. The file, “SampleID_Assay_Design.xls”, is downloadable from mysequenom.com and contains all the essential assay design specifications for the iPLEX Pro Sample ID Panel procedure. • Configure the software to indicate how samples will be identified and whether a sample is a normal or tumor sample. 3.1 Importing the Sample ID Assay Group File 1. Open Typer AssayEditor. 2. Create a new Assay Project in the Database Browser by right-clicking the root node and selecting Project Administrator. 3. Add a new Assay Project with an appropriate name. The new Assay Project will appear in the database browser. The Sample ID assay group file will be stored in this project. 4. Right-click on the newly created Assay Project and select Import Assay Group in Designer format... 5. Remove the check marks next to Design Summary and SNP Group. Make sure that there is a check mark next to Assay Group. 6. Click the Browse button next to Assay Group. 7. Navigate to folder docs under Typer 4.0 install and select “SampleID_Assay_Design.xls”. 8. Click the Import button to import the group file. 9. You can now use the imported Sample ID assays to design a Sample ID plate using PlateEditor that corresponds with your plate layout. NOTE The instructions presented here are for users who are familiar with importing assays using Typer AssayEditor and designing plates with Typer PlateEditor. For comprehensive information on using Typer, refer to the Typer 4.0.20 User’s Guide (UG 11557), which is available by contacting Sequenom Customer Support or from mysequenom.com. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 10 Chapter 3 Configuring the Software 3.2 Setting Up Sample Identification There are two important pieces of information that the user needs to provide for each sample, in addition to the sample name: • a sample identifier, which is a sequence of characters that determines whether two samples are expected to match. • an indicator whether or not a sample is a tumor sample.This is important because the algorithm for comparing a non-tumor sample with a tumor sample is different than the algorithm for comparing two non-tumor samples. Additionally, two tumor samples can never be compared with each other. NOTE The different options for specifying this information, described below, may impact the way you name your samples. The default method for specifying sample identifiers and tumor identifiers is for the user to provide a file with the identifiers listed (see page 13 and page 14 for instructions on how to construct those files). However, there are other options that may be chosen, which involve deriving the identifiers from the sample names. Instructions are given below for choosing one of these methods. Once you have made your sample identification choices, they will remain in effect indefinitely. If you wish at some point to change the way you are identifying your samples, you can navigate to the settings window again and change your choices. To configure your identification settings: 1. Open TyperAnalyzer and in the Project Explorer pane double click on the chip(s) to be compared. The chip(s) will be added to the Chip list. 2. Load the chip(s) by checking the box next to the chip name(s) in the Chip list. 3. Once the chip(s) are loaded, select File → Reports TyperAnalyzer menu bar. (See Figure 3.1.) iPLEX® Pro Sample ID Panel User Guide → Launch Sample ID in the SQNM-USG-CUS-050 Setting Up Sample Identification Figure 3.1 Launching the Sample ID Panel software The main control panel will appear (see Figure 3.2). Figure 3.2 Main control panel iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 11 12 Chapter 3 Configuring the Software 4. Click on Settings. The Settings window will appear. The left side of the window contains options for assigning identifiers to samples, and the right side contains options for determining whether or not a sample is a tumor sample. Figure 3.3 Settings window Specifying a Sample Identifier 5. Click on a method for identifying matching samples. (See page 13 for a description of these choices.) 6. Click on a method for identifying tumor vs. non-tumor samples. (See page 14 for a description of these choices.) 7. Click Save. The sample identifier is a sequence of characters that determines whether two samples are expected to match. It may be equal to the sample name, derived from the sample name, or completely different from the sample name. Consider the following three samples. If we specify that the sample identifier is the first 5 characters of the sample name, we get the following: Sample Name Sample Identifier Sample B 12345_1_N 12345_2_T Sample C 67890_1_N 12345 12345 67890 Sample A Samples A and B share a common identifier, indicating that they are expected to match. Samples A and C and samples B and C do not share common identifiers, so these are not expected to match. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Setting Up Sample Identification The choices available in the Settings window for specifying sample identifiers are described below. Options for Identifying Matching Samples in the Settings Window • A file will be provided when the Sample ID panel is run (this is the default setting) If this option is selected, each time you start a new comparison the software will ask you to specify a file that contains a list of sample names and the identifiers assigned to these samples. You may also select a default location for these files to be located. The sample name/identifier file must be a .csv file, with the following format: If a line has two columns: Column 1 = Typer sample name Column 2 = Sample identifier If a line has three columns: Column 1 = Plate ID Column 2 = Well Column 3 = Sample identifier You may mix and match different acceptable formats on different lines of the same file. • A delimited section of the sample name If this option is selected, the sample name is separated into sections based on a delimiting character, and then the nth section is chosen as the sample identifier. You must specify the character to be used (Separator) and the value of n (Section Number), which must be an integer between 1 and 30 or between -1 and -30 (negative numbers start from the last section and work backwards). See the examples below. Separator Sample Name Section Number Sample Identifier - 12345-45-67 1 12345 _ 12345_678_90 -1 90 • A character range of the sample name If this option is selected the sample identifier will be a character range of the sample name. You must specify a starting and an ending character, and the sample identifier is the range of characters between and including these characters. Each must be an integer from 1 to 60 or from -1 to -60 (negative numbers start from the last character and work backwards). See the examples below. Starting character Ending Character Sample Name Sample Identifier 1 8 ABCDEFGHIJKLMNOP ABCDEFGH 5 6 1234567890 56 -5 -1 1234567890 67890 2 -2 1234567890 23456789 iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 13 14 Chapter 3 Configuring the Software • Keep Typer sample name If this option is selected the sample identifier will be the same as the sample name defined in the Typer software. See the examples below. Sample Name Sample Identifier 12345_1 12345_2 12345_1 12345_2 67890_1 67890_1 Specifying a Tumor Identifier Each sample must be identified as either tumor or non-tumor. Consider the following three examples. In this case the last character could be used to determine whether or not a sample is a tumor sample, with the presence of a “T” indicating a tumor sample. Sample Name Tumor Sample? Sample B 12345_1_N 12345_2_T Sample C 67890_1_N No Yes No Sample A These definitions lead to the following comparison algorithms being used by the Sample ID software: Samples A and B: Non-tumor vs. tumor algorithm Samples A and C: Non-tumor vs. non-tumor algorithm Samples B and C: Non-tumor vs. tumor algorithm The choices available in the Settings window for specifying tumor identifiers are described below. Options for Identifying Tumor Samples in the Settings Window • A file will be provided when the Sample ID panel is run (this is the default setting) If this option is selected, each time you start a new comparison the software will ask you to specify a file that contains a list of sample names labeled as tumor samples. You may also select a default location for these files to be located. The tumor identifier file must be a .csv file, with the following format: If a line has one column: Column 1 = Typer Sample Name (this will be defined as a tumor sample) If a line has two columns: Column 1 = Plate ID Column 2 = Well (this well on this plate will be defined as a tumor sample) You may mix and match different acceptable formats on different lines of the same file. Any sample not present in the tumor identifier file is considered a non-tumor sample. • A delimited section of the sample name If this option is selected, the sample name is separated into sections based on a delimiting character, and if the nth section matches a designated tumor value then the sample is labeled a tumor sample. You must specify the character to be used (Separator), the Tumor Value, and the value of n (Section Number), which must be an integer between 1 and 30 or between -1 and -30 (negative numbers start from the last section and work backwards). See the examples below. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Setting Up Sample Identification Separator - Section Number 1 Tumor Value T Sample Name Tumor Sample? T-12345-ABC Yes 12345-ABC No N-12345-ABC No • Starts with the given value If this option is selected, the sample will be identified as a tumor sample if the sample name starts with a user-specified tumor value. See the examples below. Tumor Value Sample Name Tumor Sample? 12345 No T T12345 Yes 12345-T No • Ends with the given value If this option is selected, the sample will be identified as a tumor sample if the sample name ends with a user-specified tumor value. See the examples below. Tumor Value T Sample Name Tumor Sample? 12345 No T12345 No 12345-T Yes 12345T Yes • Contains the given value If this option is selected, the sample will be identified as a tumor sample if the sample name contains a user-specified tumor value in any location. See the examples below. Tumor Value T iPLEX® Pro Sample ID Panel User Guide Sample Name Tumor Sample? 12345 No T12345 Yes 12345T Yes 123T456 Yes SQNM-USG-CUS-050 15 16 Chapter 3 Configuring the Software iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 4 Assay Protocol Isolating DNA from Samples . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Performing PCR Amplification . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Performing the SAP Treatment . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Performing the iPLEX Pro Extend Reaction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Desalting the Extend Reaction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Designing a Sample ID Plate in PlateEditor . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17 17 19 20 22 23 4.1 Isolating DNA from Samples For each sample, prepare 10 ng of genomic DNA in a volume of 2 μl. ! IMPORTANT Perform this procedure in laboratory area 1. Types of Samples The iPLEX Pro Sample ID Panel can be performed on genomic DNA isolated from fresh tissue, frozen tissue, formalin-fixed paraffin-embedded (FFPE) tissue, or cell lines. Method You may use any method that you prefer to isolate genomic DNA from your samples. Note that DNA isolated from FFPE samples is usually of lower molecular weight than DNA from fresh or frozen samples. The degree of fragmentation depends on the type and age of the sample and the conditions used for fixation. A commercial, lysis-based kit such as Qiagen’s QIAamp DNA FFPE Tissue Kit or a similar product is recommended for processing FFPE samples. Purity Use a spectrophotometric or fluorometric method to determine the concentration and relative purity of your genomic DNA. Amount The amount of genomic DNA needed for the iPLEX Pro Sample ID Panel is 10 ng per sample. A maximum of 960 samples (for the 10 x 96 kit) or 768 samples (for the 2 x 384 kit) can be assayed. Storage Purified genomic DNA can be stored at 2-8 °C for up to 24 hours. To store DNA longer than 24 hours, we recommend storage at -25 to -10°C. Avoid freezing and thawing DNA. More Information For further detailed information on purification, storage, quantification. and analysis of genomic DNA, refer to the Genomic DNA Bench Guide, available at the Qiagen web site. 4.2 Performing PCR Amplification Genomic DNA from your samples will be amplified using the supplied iPLEX Pro Sample ID Panel PCR primers in either a 96-well or a 384-well plate in a final reaction volume of 5 μl. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 18 Chapter 4 Assay Protocol ! IMPORTANT Perform the PCR cocktail preparation and addition of DNA to the cocktail in the reaction plate in laboratory area 2. Preparation performing pre-PCR cycling work, clean the bench areas, pipettes, and hood work area • Before with DNA AWAY or similar solution, followed by 70% ethanol, and illuminate with UV light for 30 minutes. • Clean post-PCR bench areas with DNA AWAY. vortex and centrifuge tubes and plates that contain reagents or samples before • Briefly proceeding to the next step in the protocol. • When not in use, seal plates with adhesive PCR sealer and store at the appropriate temperature. • Plate seals are recommended to minimize evaporation: Adhesive PCR Film Cs/100 (Myriad Industries) Bio-Rad Pressure Pads (#ADR-5001) following thermocyclers are recommended due to the high likelihood of evaporation of the • The low reaction volume in 96-well plates: Perkin Elmer PE7500, PE9700 ABI: GeneAmp PCR System 9700 96-well NOTE The use of MJ Research Tetrad cyclers is strongly discouraged. • Store stock reagents and finished cocktails in plates at -20°C when not in use. • Wear gloves during the entire procedure. Procedure 1. Add the following reagents to a 1.5 ml Eppendorf tube. NOTE If using UNG enzyme during PCR, see the alternative UNG PCR protocol in Appendix B. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Performing the SAP Treatment Volume per 96-well Plate* Volume per 384-well Plate* Initial Concentration Final Concentration HPLC-grade water N/A N/A 0.3 μl 36.3 μl 145.1 μl 10X PCR Buffer 10X 1X 0.5 μl 60.5 μl 241.9 μl Reagent 1X MgCl2 25 mM 2 mM 0.4 μl 48.4 μl 193.5 μl dNTP Mix 25 mM 500 μM 0.1 μl 12.1 μl 48.4 μl iPLEX Pro Sample ID Panel PCR Primer Set 500 nM 100 nM 1.0 μl 121.0 μl 483.8 μl Sequenom PCR Enzyme 5 U/μl 1 U/rxn 0.2 μl 24.2 μl 96.8 μl 10X 1X 0.5 μl 60.5 μl 241.9 μl 3.0 μl 363.0 μl 1451.4 μl Q-Mix Final Volume *includes 26% overhang 2. Mix well by brief vortex and centrifuge briefly. 3. Dispense 3 μl of the PCR mix into each well of a 384-well plate if using a 384-plate set up OR a 96-well plate if using a 96-plate set up. 4. Dispense 2 μl of DNA into each well of the plate containing the PCR mix. Make sure that the pipette tips do not come into contact with the PCR cocktail already in the plate to avoid cross-contamination of assays. 5. Seal the plate with thermal sealing film, vortex briefly, and centrifuge. 6. Perform thermocyling using the following conditions: 95°C 2 minutes 95°C 30 seconds 56°C 30 seconds 72°C 1 minute 72°C 5 minutes 4°C Hold 45 cycles 4.3 Performing the SAP Treatment After amplification, any remaining free deoxynucleotides in the amplification reaction mixture must be dephosphorylated to prevent interference with the iPLEX Pro reaction. Shrimp alkaline phosphatase (SAP) dephosphorylates unincorporated dNTPs and converts them to dNDPs, making them unavailable for future reactions. The SAP is then heat inactivated at 85°C. ! IMPORTANT Perform the SAP addition in laboratory area 3. . iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 19 20 Chapter 4 Assay Protocol Procedure 1. Prepare the following SAP mix in a 1.5 ml Eppendorf tube: Volume per 96-well Plate* Volume per 384-well Plate* Initial Concentration Final Concentration HPLC-grade water n/a n/a 1.53 μl 198.3 μl 793.2 μl 10X SAP Buffer 10X 1X 0.17 μl 22.0 μl 88.1 μl SAP (1.7 U/μl) 1.7 U/μl 0.07 U/μl 0.30 μl 38.9 μl 155.5 μl 259.2 μl 1036.8 μl Reagent 1X Final Volume *includes 35% overhang 2. Dispense 2 μl of SAP mixture to each well of the reaction plate containing the PCR products. You can also use the SAP addition protocol on the Sequenom Liquid Handler. 3. Seal the plate with thermosealing film, vortex plate briefly to mix, and centrifuge plate. 4. Perform thermocycling using the following conditions: 37°C 40 minutes 85°C 5 minutes 4°C Hold 1 cycle 4.4 Performing the iPLEX Pro Extend Reaction The iPLEX Pro Sample ID Panel extend reaction is a method for determining sample identification and genomic copy number quantification. The iPLEX Pro Extend reaction cocktail is added to the amplification products produced in the previous step. The amplification products and reaction cocktail are thermocycled, allowing the enzymatic addition of a nucleotide into the diagnostic site. The primer is extended by one nucleotide, terminating the primer extension. The iPLEX Pro Extend reaction produces allele-specific extension products of different masses depending on the sequence analyzed (Table 4.1). Table 4.1 iPLEX Pro Extend Reaction Products Required Materials Analytes Peak Description Length of Product (bp) Calculated Mass (Da) Unextended primer Extension primer 20 6163.0 Low mass allele Extension Primer + A 21 6434.2 High mass allele Extension Primer + G 21 6450.2 You will need the following materials on hand before beginning: • SAP-treated PCR samples in 384 plate(s) OR 96 plate(s) • Sequenom iPLEX Pro Reagent Set • iPLEX Pro Sample ID Extend primer, thawed iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Performing the iPLEX Pro Extend Reaction • 1 full-skirted 96-well PCR plate • PCR sealing film • RNAse and DNAase-free water • 12-multichannel pipettors and sterile pipette tips • Ice bucket with ice Procedure ! IMPORTANT Perform the extend reaction in laboratory area 3. 1. Prepare the iPLEX Pro Extend reaction mix on ice in a 1.5 ml Eppendorf tube as follows: Reagent Initial Concentration Final Concentration 1X Volume per 96-well Plate* Volume per 384-well Plate* HPLC-grade water n/a n/a 0.755 μl 97.8 μl 391.4 μl iPLEX Pro Buffer Plus (10X) 10X 0.222X 0.200 μl 25.9 μl 103.7 μl Thermosequenase Termination Mix 10X 0.222X 0.200 μl 25.9 μl 103.7 μl Extension Primer Mix n/a n/a 0.804 μl 104.2 μl 416.8 μl 32 U/μl 0.15 U/μl 0.041 μl 5.3 μl 21.3 μl 2.000 μl 259.1 μl 1036.9 μl ThermoSequenase (32 U/μl) Final Volume *includes 35% overhang 2. Dispense 2 μl of the Extend reaction mix into each well of the SAP-treated PCR reaction plate. Make sure the tips of the multichannel do not come into contact with the PCR product already in the plate to avoid assay cross-contamination. 3. Seal the plate and vortex briefly to mix. 4. Centrifuge at 4,000 rpm for 5 seconds. 5. Thermocycle the plate using the following reaction conditions: 95°C 30 seconds 95°C 5 seconds 52°C 5 seconds 80°C 5 seconds 72°C 3 minutes 4°C Hold iPLEX® Pro Sample ID Panel User Guide 5 cycles 40 cycles SQNM-USG-CUS-050 21 22 Chapter 4 Assay Protocol 4.5 Desalting the Extend Reaction Required Materials You will need the following materials on hand before beginning: • CLEAN Resin • 384/6 mg dimple plate for 384-plate set up OR 96/15 mg dimple plate for 96-plate set up • iPLEX Pro Extend reaction plate from previous procedure • PCR sealing film • HPLC-grade water • 12-multichannel pipettors and barrier filter pipette tips • Plate rotator (at room temperature) Procedure ! IMPORTANT Wear gloves and safety glasses during the desalting procedure. 1. Spread CLEAN Resin on a clean, dry dimple plate (~3 scoops per plate) using the scraper plate. Make sure that the resin settles evenly into all wells. 2. Let the resin plates dry for 10 minutes at room temperature. 3. While the resin plates are drying, add 16 μl (for 384-well plate setup) OR 41 μl (for 96-well plate set up) HPLC-grade water to each well of the iPLEX Pro Extend reaction product plates using a 12-channel multipipettor. 4. To add dried CLEAN Resin to each well: a. Gently invert the sample plate on top of the dimple plate, making sure that the plate wells are aligned over the resin samples. b. Keep the sample and dimple plates pressed together, and invert both plates so that the dimple plate is on top of the sample plate. c. Gently tap the dimple plate and let the resin fall into the sample wells. 5. Check the plate to make sure that the resin height is the same in all wells. Make note of any wells that have more resin. 6. Seal the plate and rotate for 30 minutes at room temperature. The rotator must rotate the microplate 360° around its long axis. 7. Centrifuge the plate at ~3,200 g for 5 minutes to pellet the resin. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Designing a Sample ID Plate in PlateEditor 4.6 Designing a Sample ID Plate in PlateEditor The Sample ID plate layout and sample specifications have to be designed in PlateEditor in order to acquire data on the MassARRAY system and to perform data analysis in Typer. Design a Sample ID plate using PlateEditor that corresponds with your plate layout. See page 10 for information on choosing sample names. NOTE For comprehensive information on using Typer, refer to the Typer 4.0.20 User’s Guide (UG 11557), which is available by contacting Sequenom Customer Support or from mysequenom.com. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 23 24 Chapter 4 Assay Protocol iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 5 Dispensing Samples to a SpectroCHIP® Array Preparing the Nanodispenser and Reaction Plate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Selecting Nanodispenser Settings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Loading the Reaction Plate and SpectroCHIP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Starting the Dispensing Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Removing the SpectroCHIP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25 26 26 26 27 You will dispense the samples from the conditioned reaction plate onto a SpectroCHIP array using the RS1000 Nanodispenser. The main steps to dispense samples from the microplate onto the array include: • Preparing the Nanodispenser and reaction plate. • Selecting Nanodispenser settings. • Loading the reaction plate and SpectroCHIP. • Starting the dispensing run. • Removing the SpectroCHIP after the run is completed. ! IMPORTANT The iPLEX Pro Sample ID Panel was validated using a MassARRAY Nanodispenser RS1000. For further operating information, see the appropriate MassARRAY Nanodispenser User’s Guide. NOTE Before dispensing samples, confirm that the daily and weekly instrument maintenance procedures have been performed. NOTE The instructions presented here are for users who are familiar with dispensing samples using a MassARRAY Nanodispenser. For comprehensive instructions refer to the MassARRAY Nanodispenser RS1000 User’s Guide, which is available by contacting Sequenom Customer Support or from mysequenom.com. 5.1 Preparing the Nanodispenser and Reaction Plate 1. Turn on the Nanodispenser and log on. 2. Check the supply and waste tanks, and the ultrasonic wash supply bottle. • A full supply tank is enough to process ten 384-well SpectroCHIPs. • An empty waste tank has enough capacity to process ten 384-well SpectroCHIPS. • A full bottle of ultrasonic wash supplies enough solution for a day of instrument use. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 26 Chapter 5 Dispensing Samples to a SpectroCHIP® Array Fill the calibrant reservoir with 80 μl of 3-point calibrant. 3. ! CAUTION If there is calibrant in the reservoir that will not be used within a few hours, remove the calibrant with a pipette and return it to the calibrant bottle for storage. 4. Check to make sure that the resin levels and liquid are uniform across the microplate wells. The liquid level must be at least 2.5 mm above the resin. 5. Record the instrument, SpectroCHIP barcode, and dispensing parameters for your records. 5.2 Selecting Nanodispenser Settings 1. Park the pin array assembly. 2. On the Main Menu screen, tap Transfer. On the Transfer screen, tap Open and select the appropriate method file, either “96 Plate to 96 Chip” or “384 Plate to 384 Chip.” 3. On the Transfer screen, tap the Methods button, and select the aspirate/dispense tab. “dispense settings” enter a dispense speed of approximately 80-120 mm/sec, • Under depending upon environmental conditions. The target volume for dispensing iPLEX Pro Sample ID Panel assays is 15 nl. • Under “calibrant” enter a dispense speed of at least 140 mm/sec. • Under “operation” choose analyte & calibrant. 4. On the Method screen, select the setup tab, and under “Analysis” select volume check. 5.3 Loading the Reaction Plate and SpectroCHIP 1. Place the 96- or 384-well reaction plate on plateholder 1, such that well A01 is to the lower left. Be sure to turn the hold-down latches so they clip onto the top of the plate. 2. Remove the SCOUT plate or Compact plate adapter from the processing deck and insert a SpectroCHIP into the plate in the chip position you selected earlier. 3. Place the SCOUT plate or Compact plate adapter back onto the processing deck. 4. Close the main door. 5. In the safety interlock is disengaged! dialog box, tap the HOME button. 5.4 Starting the Dispensing Run 1. On the Transfer screen, tap the step icon to start the dispensing run. 2. When the instrument pauses, tap the volume button to check droplet volume. If necessary, adjust the dispense speed to get to a target volume of 15 nl per droplet. 3. Disable volume check, and then continue the run by tapping the run icon on the Transfer screen. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Removing the SpectroCHIP 5.5 Removing the SpectroCHIP 1. Park the pin array assembly and transfer the chip to the appropriate chip holder for analysis in the MassARRAY Analyzer. NOTE It is recommended that you process the SpectroCHIP array immediately on the MassARRAY Analyzer. However, if you are not able to process the chip immediately, return it to the protective clamshell case and store in a desiccator with fresh desiccant away from ambient light. \ 2. Remove the sample microplate, seal, and store at -20°C if you want to save the plate for future analysis. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 27 28 Chapter 5 Dispensing Samples to a SpectroCHIP® Array iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 6 Acquiring Data Starting the MassARRAY Analyzer Software . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Loading the SpectroCHIP Array(s) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Creating an Input File using Typer Chip Linker . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Setting Up the Automatic Run and Checking the Barcode Report . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Starting the Automatic Run . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Unloading the SpectroCHIP Array(s) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29 29 30 31 32 33 The main steps in acquiring data from the MassARRAY Analyzer are: • Start the MassARRAY Analyzer software • Load the SpectroCHIP array(s) • Create an input file using Typer Chip Linker software • Set up the automatic run and check the barcode report • Start the automatic run • Unload the SpectroCHIP array(s) NOTE The instructions presented here are for users familiar with analysis using the MassARRAY Analyzer 4. For more information on how to operate the MassARRAY Analyzer, or for MassARRAY Analyzer Compact instructions, refer to the appropriate user’s guide [MassARRAY Analyzer 4 User’s Guide (UG 161099) or MassARRAY Analyzer Compact User’s Guide (UG 11533)], available from Sequenom Customer Support or from mysequenom.com. 6.1 Starting the MassARRAY Analyzer Software To launch the MassARRAY Analyzer 4 application, double-click the Analyzer 4 on the desktop. instrument icon To launch the MassARRAY Analyzer Compact application, double-click the Start RT Processes icon on the desktop. 6.2 Loading the SpectroCHIP Array(s) 1. To load a SpectroCHIP array into the chip holder, or to place a SpectroCHIP arraycontaining chip holder on the chip carrier/target, probe the chip carrier out by pressing the appropriate manual control button on the front of the MassARRAY Analyzer, or by clicking Probe Sample In/Out or Target In/Out on the software toolbar. It takes approximately one minute for the chip holder to move to the load position. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 30 Chapter 6 Acquiring Data ! CAUTION When the chip holder is moving in or out of the analyzer, do not start or quit data acquisition. Do not move the chip holder during spectra acquisition. Wait until acquisition is completed, or abort the run. 2. Open the sample chamber lid and take out the chip holder. Remove the previously run chips from the chip holder. 3. Place the chips to be processed into the chip holder. Orient the SpectroCHIP array in the chip holder so that the Sequenom logo is at the bottom. Make sure the lower left corner of the SpectroCHIP array is flush against the chip holder. Chip 1 is seated on the left and chip 2 is seated on the right. If there is only one chip with samples, it is recommended that you place a second dummy chip in the chip holder for better results. Also make sure that the samples are dried and no liquid or dust is on the chip before loading. NOTE Always wear gloves and use the supplied tweezers when handling SpectroCHIPs. 4. Close the sample chamber lid and retract the target by pressing the manual control button on the instrument, or selecting Target In/Out or Probe Sample In/Out in the software tool bar. The chip holder moves to the “sample in” position. The analyzer ready light illuminates when the required vacuum pressure (5 x 10 -6) is reached. This takes approximately two minutes. ! IMPORTANT Only use the chip holder that was supplied with the MassARRAY Analyzer. The chip holder design is instrument-specific and chip holders are not interchangeable between instruments. 6.3 Creating an Input File using Typer Chip Linker An input file provides the information that SpectroACQUIRE needs to process a chip and save data to the MassARRAY database. 1. Double-click the Chip Linker icon 2. In the dialog box that appears, enter your user name, password, and server. 3. Click Connect or OK. The Chip Linker window appears. See Figure 6.1. 4. In the Chip Linker directory tree, browse to your experimental folder and select your plate that was created in Plate Editor. 5. Select the iPLEX Terminator Chemistry button. 6. Select "genotype+area" for the process method. 7. For Dispenser, select Nanodispenser 384 to 384 OR Nanodispenser 96 to 96 as appropriate. iPLEX® Pro Sample ID Panel User Guide on the workstation desktop. SQNM-USG-CUS-050 Setting Up the Automatic Run and Checking the Barcode Report 8. Enter an experiment name. When entering the information, use alphanumeric characters excluding special characters such as \/:*?<>|, and spaces. 9. For Chip Barcode, enter the SpectroCHIP barcode located on the chip. 10. Click Add. The input information appears in the Chip Linker table. 11. If a second chip will be processed, repeat step 4 to step 10 for the second chip. 12. Click Create to create an input .xml file. This file will be selected for use when you set up the automatic run. Close the Typer Chip Linker software. Figure 6.1 Chip Linker window ! IMPORTANT In the Chip Linker table, "Index 1” refers to chip holder position 1 and “Index 2” refers to chip holder position 2. 6.4 Setting Up the Automatic Run and Checking the Barcode Report 1. In SpectroACQUIRE, click the Automatic Run Setup tab. 2. Enter Acquisition Parameters and Geometry settings as shown in Figure 6.2. 3. In the Chip 1 box, enter the barcode that was entered in Chip Linker. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 31 32 Chapter 6 Acquiring Data NOTE The chip number in the Automatic Run Setup tab corresponds to the chip position in the chip holder. Chip 1 is seated on the left and chip 2 is seated on the right. 4. Click Barcode Report. In the dialog box that appears, each chip position should have a FOUND status. This means that the SpectroCHIP is properly associated with an experiment in the MassARRAY database. 5. Click CLOSE and correct any chip positions with an Error status. Figure 6.2 SpectroACQUIRE Automatic Run Setup Tab 6.5 Starting the Automatic Run 1. In SpectroACQUIRE, click the Automatic Run Setup tab. 2. To start the run, click the button on the SpectroACQUIRE toolbar or select Run → Start Auto Run on the menu bar. The software: ! • Checks the name of the SpectroCHIP array and confirms that the name is associated with an experiment in the MassARRAY database. • Checks the position of the chip in the chip holder and performs autoteaching. • Validates SpectroCHIP 2D barcode. • Acquires calibration spectra. • Acquires spectra. IMPORTANT The software checks the name of each SpectroCHIP array when the SpectroACQUIRE software begins to process it. If the software encounters an error in a name, the run stops with that particular SpectroCHIP array. Click Barcode Report (on the Automatic Run Setup tab) to check all SpectroCHIP names before starting a run. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Unloading the SpectroCHIP Array(s) 6.6 Unloading the SpectroCHIP Array(s) Do not unload SpectroCHIP arrays while SpectroACQUIRE is acquiring spectra. Wait until the data acquisition is completed. 1. In the SpectroACQUIRE software, click the Automatic Run Setup tab. 2. Press the Probe Sample In/Out or Target In/Out button on the front of the Analyzer to extend the target. It takes approximately one minute for the chip holder to move to the load position. ! CAUTION When the chip holder is moving in or out of the analyzer, do not start or quit the SpectroACQUIRE software or start acquisition. Do not move the chip holder during spectra acquisition. Wait until acquisition is completed, or abort the run. 3. Open the sample chamber lid and take out the chip holder. Remove the chips from the chip holder. Replace chip holder onto the chip carrier. Click the Probe Sample In/Out or Target In/Out button on the front of the Analyzer to retract the chip carrier. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 33 34 Chapter 6 Acquiring Data iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Chapter 7 Analyzing Data Generating Sample ID Reports . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35 Reading Sample ID Reports . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 39 This section contains instructions for generating the Sample ID reports, and descriptions of the Sample ID report files. NOTE The instructions presented here are for users who are familiar with analyzing data with Typer. For comprehensive information on using Typer, refer to the Typer 4.0.20 User’s Guide (UG 11557), which is available by contacting Sequenom Customer Support or from mysequenom.com. 7.1 Generating Sample ID Reports 1. Open TyperAnalyzer and in the Project Explorer pane double click on the chip(s) to be compared. The chip(s) will be added to the Chip list. 2. Load the chip(s) by checking the box next to the chip name(s) in the Chip list. 3. Once the chip(s) are loaded, select File → Reports → Launch Sample ID in the TyperAnalyzer menu bar. (See Figure 7.1.) Figure 7.1 Generating Sample ID Reports iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 36 Chapter 7 Analyzing Data NOTE Only one chip per plate may be loaded in the Sample ID software. If you load two chips from the same plate, an error message will appear. The main control panel will appear (see Figure 7.2). At the top of the window the plates that are currently loaded in Typer are displayed. The Stored column indicates whether or not these plates are currently stored in the Sample ID database. Figure 7.2 Main control panel From the main control panel you may generate reports, delete loaded plates from the Sample ID database, access the Sample ID settings window, or quit the Sample ID software (see Table 7.1). iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Generating Sample ID Reports Table 7.1 Functions in Main Control Panel Button Function Historical Performs a Sample ID comparison of the loaded plates against themselves and against the entire Sample ID database. Data from these plates will be stored in the Sample ID database and a report will be generated. Local Performs a Sample ID comparison of the loaded plates against themselves. This option does not query the Sample ID database and will not store information about these plates in the Sample ID database. A report will be generated. Delete Deletes the loaded plates from the Sample ID database. This has no effect on the data stored in the Typer database, and does nothing if the loaded plates are not currently stored in the Sample ID database. Settings Opens the Sample ID settings window (see page 10 for more information). Quit Quits the Sample ID panel software and returns you to TyperAnalyzer. NOTE If you load a chip from a plate that already has another chip stored in the database, a notice will appear in the main control panel, because you may only store one chip per plate in the Sample ID database. You may run a local comparison on the loaded chip, and this will use data from the loaded chip only, as the database is not used for a local comparison. You may run a historical comparison, but data from the currently loaded chip will be ignored and data from the chip previously stored in the database will be used. 4. To generate a report, click on either Historical or Local in the main control panel. The software will indicate what type of comparison is being done (“Performing comparison only within loaded plates” in Figure 7.3 indicates that Local was selected.). iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 37 38 Chapter 7 Analyzing Data Figure 7.3 Sample ID Report being generated 5. When the comparison is complete, results will be made available in two different formats (.csv and HTML) and stored in the Typer\bin folder, as indicated in the main control panel (see Figure 7.4). The HTML index file (the Summary Report) will automatically open in your browser. Figure 7.4 Sample ID Report completed iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Reading Sample ID Reports NOTE If the HTML reports do not open properly, make sure you are using a compatible browser, i.e., Internet Explorer or Chrome. 7.2 Reading Sample ID Reports The two .csv files contain all the comparison and sample data generated; the four HTML files contain data in a more readable format (see Table 7.2). Table 7.2 Report Files Generated File type Folder or file name .csv matches This file contains information on all of the comparisons from the three reported categories (unexpected mismatches, unexpected matches, and expected matches). It contains information about the two samples involved in the result, the score of the match, whether or not it was a match or a mismatch, and whether or not it was expected. samples This file contains information on all of the samples from the loaded plate(s), including information about the SNP calls, the number of amplifiable copies, the gender, and the quality control results. matches This folder contains files for each unexpected match and mismatch and each expected match; each match file includes information for the two samples involved in the comparison, details of the SNP calls for each sample, and the match scoring. plates This folder contains files for each of the loaded plates. Each plate file contains results for each well on that plate, including number of SNP calls, number of amplifiable copies, gender, whether it passed quality control, and the number of unexpected matches and mismatches and expected matches found. samples This folder contains files for each sample in the loaded plates. Each sample file contains copy number, gender, and SNP assay results for that sample, as well as details of any unexpected matches or mismatches or expected matches of that sample with any other sample. If the sample failed quality control the reason(s) will be stated. index This file contains the Summary Report, including a list of unexpected matches and mismatches and information on samples that did not pass quality control. This file will automatically open in your browser once the report files have been generated. HTML HTML files Contents Upon completing a Sample ID comparison, the HTML-formatted Summary Report will automatically open in your browser window. (See Figure 7.5.) The Plate Report, Match Report, and Sample Report may all be accessed from the Summary Report. Note that expected matches are not listed in the Summary Report, but are included in the Plate, Match, and Sample Reports. Summary Report The Summary Report consists of four sections, as shown in Figure 7.5: • Report identifying information, including type of report, date, plates compared, etc. Clicking on the plate ID name will take you to the Plate Report. • Summary results, showing how many samples failed quality control and how many unexpected results were generated in each comparison category (non-tumor vs. non-tumor mismatches, non-tumor vs. non-tumor matches, non-tumor vs. tumor mismatches, and non-tumor vs. tumor matches). Clicking on the number of failed samples will take you to the QC Summary section farther down the Summary Report; clicking on the number of unexpected results will take you to the more detailed unexpected results section farther down the Summary Report. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 39 40 Chapter 7 Analyzing Data • Unexpected results details, including identifying information for each of the samples that were compared, as well as a match score (see Table 7.3). Low confidence match scores are indicated by a yellow background. Clicking on the match score will take you to the Match Report. Table 7.3 Match scores Non-tumor vs. non-tumor comparisons Non-tumor vs. tumor comparisons Match 0 to 10; 10 is a perfect match Mismatch <0 Low confidence score -5 to 5 Match 0 to 5; 5 is a perfect match Mismatch <0 Low confidence score -3 to 3 • Quality control (QC) details, lists all samples that failed quality control, and gives the reason(s). Clicking on the sample name will take you to the Sample Report. Samples may fail quality control for the following reasons: -Sample contains fewer than the minimum allowable number of amplifiable copies (500). -Sample contains fewer than the minimum allowable number of SNP calls (30). -Sample contains assays that are not a part of the Sample ID Panel. -Sample is missing assays that are a required part of the Sample ID Panel. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Reading Sample ID Reports Identifying information Click to see Plate Report. Click to move to QC Summary below. Click to move to unexpected matches list below. Summary results Click to see Match Report. Unexpected matches/ mismatches detail Click to see Sample Report. Quality control results details Figure 7.5 Sample ID Report Summary iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 41 42 Chapter 7 Analyzing Data Plate Report There is a separate HTML file for each plate that was loaded. The Plate Report lists results for every well in the plate, including number of SNP calls, number of amplifiable copies, gender, whether it passed quality control, and the number of matches and mismatches found. Clicking on the sample name will take you to the Sample Report, where you can see details on unexpected matches and mismatches and expected matches for that sample. Click here to access Sample Report Figure 7.6 Plate Report Sample Report There is a separate HTML file for each sample on the loaded plate(s). Each Sample Report contains results of the five copy number assays, the three gender assays, and the 44 SNP assays. It also reports the details of each unexpected match or mismatch and each expected match with other samples; clicking on the match score will take you to the Match Report for that comparison (see Figure 7.7). If the sample failed quality control, the Sample Report will list the reason(s) (see Figure 7.8). The legend at the end of the report shows how SNP calls are categorized as aggressive, moderate, or conservative, and how SNP matches and mismatches are color-coded. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Reading Sample ID Reports Click here to see the Sample Report for this sample. Click here to see the Match Report for this comparison. Figure 7.7 Sample Report iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 43 44 Chapter 7 Analyzing Data Figure 7.8 Sample report for a failed sample iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Reading Sample ID Reports Match Report There is a separate HTML file for each unexpected match or mismatch and each expected match. The Match Report lists identifying information for the two samples involved in the comparison, and details of the SNP calls for each sample and the match scoring (see Figure 7.9). When two samples are compared, each of the 44 pairs of SNP calls are compared individually. If either sample generated a no call, that SNP is ignored for that comparison. If a non-tumor and a tumor sample are being compared (see Figure 7.10), only SNPs that are homozygous for the nontumor sample are considered, to account for potential loss of heterozygosity in the tumor. If the two SNP calls do not match, an assay-specific penalty is applied to the comparison score between those two samples, with homozygote to homozygote mismatches penalized more heavily than homozygote to heterozygote mismatches. Figure 7.9 Match Report for an Unexpected Mismatch iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 45 46 Chapter 7 Analyzing Data Figure 7.10 Match Report for an Expected Match .csv Files All the data from the samples and comparisons are contained in two .csv files, samples and matches, which are saved in the Typer\bin folder location stated in the main control panel once the report generation is complete. Note that the samples.csv file will always only contain information on the samples from the loaded plates. If a historical comparison was done, however, the matches.csv file may also contain information on samples from non-loaded plates that are in the Sample ID database. Samples.csv File The samples.csv file contains data from all the samples from the loaded plates, including the SNP calls, the number of amplifiable copies, the gender, and the quality control results for each sample. See Table 7.4 for a description of all the columns in the samples.csv file. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Reading Sample ID Reports Table 7.4 Columns in the samples.csv file Column Name Description Customer The Customer value from Typer. Project The Project value from Typer. Plate The Plate value from Typer. SAMPLE_ID_PANEL_RE SULT_PK A unique identifier for the sample. This can be used to cross-reference with the matches.csv output file. Date The date of the report. Experiment The Experiment value from Typer. Gender 1/Gender 2/ Gender 3 The gender as determined by the individual gender assays (M = Male, F = Female, 0 = No Call). SIDv1_SNP01 to SID_v1_SNP44 The genotype calls of the 44 SNP assays. PLATE_PK A unique identifier for the plate. Missing Assays QC flag for detecting missing assays. Superfluous Assays QC flag for detecting superfluous assays. DNA Amount QC flag for the number of amplifiable copies of DNA. Num SNP Calls QC flag for the minimum number of SNP calls. SIDv1_SNP01_quality to SIDv1_SNP44_quality The quality values for the individual SNP calls. These match Typer quality values with the exception of "i", which indicates a low intensity call (Intensity < 1). Amplifiable Copies 1/2/3/4/5 The number of amplifiable copies as determined by the individual quantitative assays. A value of -1 indicates a failed assay. SAMPLE_PK A unique sample identifier with the Typer database. Sample Identifier The sample identifier used to link samples and determine expected matches. Passed QC Whether or not the sample passed the QC checks. Tumor Whether or not the sample is a tumor sample. User The Typer user. Version The version of the Sample ID panel. Well The well. Typer Sample ID The Typer sample name. Amplifiable Copies (Avg) The average number of amplifiable copies. Amplifiable Copies (SD) The standard deviation across the 5 assays. Gender The overall gender determined from the individual gender assays. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 47 48 Chapter 7 Analyzing Data Matches.csv File The matches.csv file contains information about all of the matches from the three reported categories (unexpected mismatches, unexpected matches, and expected matches), including information about the two samples involved in the result, the score of the match, whether or not it was a match or a mismatch and whether or not it was expected. See Table 7.5 for a description of all the columns in the matches.csv file. Table 7.5 Columns in the matches.csv file Column Name Description Plate1 The plate of the first sample. Plate2 The plate of the second sample. Plate1 Date The date of the plate from the first sample. Plate2 Date The date of the plate from the second sample. Sample1 Name The name of the first sample. Sample2 Name The name of the second sample. Sample1 Identifier The identifier of the first sample used to link samples. Sample2 Identifier The identifier of the second sample used to link samples. Sample1 Well The well of the first sample. Sample2 Well The well of the second sample. SAMPLE_ID_PANEL_ COMPARISON_PK A unique value for this match. SAMPLE_ID_PANEL_ RESULT_PK1 A unique value for the first sample. Can be used to cross-reference with SAMPLE_ID_PANEL_RESULT_PK from the samples.csv file. Note: This value may reference a sample that was not in the loaded plates and thus is not present in the samples.csv file. SAMPLE_ID_PANEL_ RESULT_PK2 A unique value for the first sample. Can be used to cross-reference with SAMPLE_ID_PANEL_RESULT_PK from the samples.csv file. Note: This value may reference a sample that was not in the loaded plates and thus is not present in the samples.csv file. Score The match score. Homozygous Mismatches The number of homozygous to homozygous mismatches in the comparison. Heterozygous Mismatches The number of homozygous to heterozygous mismatches in the comparison. Homozygous Matches The number of homozygous to homozygous matches in the comparison. Heterozygous Matches The number of heterozygous to heterozygous matches in the comparison. Match Type Whether or not it is a match or a mismatch. Expectation Whether or not the result is expected. Date The date of the report. User The Typer user. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Appendix A Assay Information Table A.1 Sequenom iPLEX Pro Sample ID Panel Assay Information Assay Name Marker Name Location Function SIDv1_QC01 albumin chr4 DNA copy number assay SIDv1_QC02 albumin chr4 DNA copy number assay SIDv1_QC03 albumin chr4 DNA copy number assay SIDv1_QC04 albumin chr4 DNA copy number assay SIDv1_QC05 albumin chr4 DNA copy number assay SIDv1_XY01 AMEL_XY chrX/Y Gender assay SIDv1_XY02 ARSD_XY chrX/Y Gender assay SIDv1_XY03 TGIF2L_XY chrX/Y Gender assay SIDv1_SNP01 rs1005533 chr20 Genotyping SNP assay SIDv1_SNP02 rs1024116 chr18 Genotyping SNP assay SIDv1_SNP03 rs1028528 chr22 Genotyping SNP assay SIDv1_SNP04 rs10495407 chr1 Genotyping SNP assay SIDv1_SNP05 rs10771010 chr12 Genotyping SNP assay SIDv1_SNP06 rs11781516 chr8 Genotyping SNP assay SIDv1_SNP07 rs13050660 chr21 Genotyping SNP assay SIDv1_SNP08 rs1335873 chr13 Genotyping SNP assay SIDv1_SNP09 rs1357617 chr3 Genotyping SNP assay SIDv1_SNP10 rs1360288 chr9 Genotyping SNP assay SIDv1_SNP11 rs136337 chr22 Genotyping SNP assay SIDv1_SNP12 rs1382387 chr16 Genotyping SNP assay SIDv1_SNP13 rs1413212 chr1 Genotyping SNP assay SIDv1_SNP14 rs1454361 chr14 Genotyping SNP assay SIDv1_SNP15 rs1463729 chr9 Genotyping SNP assay SIDv1_SNP16 rs1468118 chr17 Genotyping SNP assay SIDv1_SNP17 rs1493232 chr18 Genotyping SNP assay SIDv1_SNP18 rs1982986 chr1 Genotyping SNP assay SIDv1_SNP19 rs1994997 chr12 Genotyping SNP assay SIDv1_SNP20 rs2010253 chr17 Genotyping SNP assay SIDv1_SNP21 rs2040411 chr22 Genotyping SNP assay SIDv1_SNP22 rs2046361 chr4 Genotyping SNP assay SIDv1_SNP23 rs2056277 chr8 Genotyping SNP assay SIDv1_SNP24 rs2076848 chr11 Genotyping SNP assay SIDv1_SNP25 rs214054 chr6 Genotyping SNP assay SIDv1_SNP26 rs2247221 chr18 Genotyping SNP assay SIDv1_SNP27 rs2518968 chr15 Genotyping SNP assay SIDv1_SNP28 rs251934 chr5 Genotyping SNP assay SIDv1_SNP29 rs2714854 chr7 Genotyping SNP assay SIDv1_SNP30 rs2831700 chr21 Genotyping SNP assay iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 50 Appendix A Assay Information Table A.1 Sequenom iPLEX Pro Sample ID Panel Assay Information Assay Name Marker Name Location Function SIDv1_SNP31 rs354439 chr13 Genotyping SNP assay SIDv1_SNP32 rs3819854 chr3 Genotyping SNP assay SIDv1_SNP33 rs717302 chr5 Genotyping SNP assay SIDv1_SNP34 rs727811 chr6 Genotyping SNP assay SIDv1_SNP35 rs729172 chr16 Genotyping SNP assay SIDv1_SNP36 rs740910 chr17 Genotyping SNP assay SIDv1_SNP37 rs8037429 chr15 Genotyping SNP assay SIDv1_SNP38 rs826472 chr10 Genotyping SNP assay SIDv1_SNP39 rs876724 chr2 Genotyping SNP assay SIDv1_SNP40 rs891700 chr1 Genotyping SNP assay SIDv1_SNP41 rs901398 chr11 Genotyping SNP assay SIDv1_SNP42 rs914165 chr21 Genotyping SNP assay SIDv1_SNP43 rs9583190 chr13 Genotyping SNP assay SIDv1_SNP44 rs964681 chr10 Genotyping SNP assay iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Appendix B UNG PCR Protocol If you choose to use UNG enzyme during PCR, substitute the following protocol for the standard one on page 17. 1. Add the following reagents to a 1.5 ml Eppendorf tube. Volume per 96-well Plate* Volume per 384-well Plate* Initial Concentration Final Concentration 1X HPLC-grade water N/A N/A 0.175 μl 21.2 μl 84.7 μl 10X PCR Buffer 10X 1X 0.500 μl 60.5 μl 241.9 μl MgCl2 25 mM 2 mM 0.400 μl 48.4 μl 193.5 μl dUTP/dNTP Mix 25 mM 500 μM 0.100 μl 12.1 μl 48.4 μl UNG Enzyme 5 U/μl 0.625 U/rxn 0.125 μl 15.1 μl 60.5 μl iPLEX Pro Sample ID Panel PCR Primer Set 500 nM 100 nM 1.000 μl 121.0 μl 483.8 μl Sequenom PCR Enzyme 5 U/μl 1 U/rxn 0.200 μl 24.2 μl 96.8 μl 10X 1X 0.500 μl 60.5 μl 241.9 μl 3.000 μl 363.0 μl 1451.5 μl Reagent Q-Mix Final Volume *includes 26% overhang 2. Mix well by brief vortex and centrifuge briefly. 3. Dispense 3 μl of each of the PCR mix into the wells of a 384-well plate if using a 384plate set up OR a 96-well plate if using a 96-plate set up. 4. Dispense 2 μl of DNA into the wells of the plate containing the PCR mix. Make sure that the multichannel pipette tips do not come into contact with the PCR cocktail already in the plate to avoid cross-contamination of assays. 5. Seal the plate with thermal sealing film, vortex briefly, and centrifuge. 6. Perform thermocyling using the following conditions: 30°C 10 minutes 95°C 2 minutes 95°C 30 seconds 56°C 30 seconds 72°C 1 minute 72°C 5 minutes 4°C Hold iPLEX® Pro Sample ID Panel User Guide 45 cycles SQNM-USG-CUS-050 52 Appendix B UNG PCR Protocol iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Appendix C Troubleshooting Table C.1 Troubleshooting Problem Possible Reason Possible Solution Too many unexpected matches or mismatches. Improper sample naming. Rename samples using nomenclature as described on page 10. Chips are over or under-dispensed. Check the spotting and perform a volume check to make sure correct volumes are dispensed on the chip. Redispense if necessary. Too many samples failed analysis. Input DNA amount for sample(s) is too low. Repeat sample run with at least 500 amplifiable copies. Copy number is higher than expected. Reagents may be degraded. Repeat biochemistry with new reagents. Copy number is lower than expected. Sample DNA is degraded. Frequently observed with formalinfixed paraffin embedded tissue; may repeat extraction. Copy number assays show a large variation amongst one another within a sample. This observation is part of the panel design; variation is due to high multiplexed assays. Software provides an average of the 5 markers for relative quantification. Peak intensities are too low. Laser energy is too low. Before running the chip, fire on one of the calibrant pads and check that the highest calibrant peak is 150 +/- 25%. Chips are over or under-dispensed. Check the spotting and perform a volume check to make sure correct volumes are dispensed on the chip. Redispense if necessary. Peak shape is too wide. MassARRAY Analyzer may need calibration. Contact SEQUENOM Customer Support. Non template control wells have genotype calls. PCR contamination. Pre-PCR preparations must be performed in a UV-sterilized chamber; review laboratory practices. Self-extension of extend primers. Can occur in wells with no DNA. If self-extension is observed in wells with DNA, then the assay may need to be repeated. Peaks are slightly off and not aligned to the expected calls. MassARRAY Analyzer may need calibration. Contact SEQUENOM Customer Support. No genotype calls, with the presence of unextended assay peaks. iPLEX Pro Reagent Set enzyme, terminator mix, or buffer may be missing from the master mix, or the dNTP mix may be degraded. Repeat biochemistry. Poor sample quality. Repeat DNA extraction. No genotyping calls, with no unextended assay peaks. Extension primer mix missing in the master mix. Repeat biochemistry. No genotyping calls, with poor quality spectra for some wells. Chips are over or under-dispensed. Check the liquid level of wells on the sample plate; check the spotting and perform a volume check to make sure correct volumes are dispensed on the chip. Re-dispense if necessary. No genotyping calls, with peaks present but not at the expected masses. Wrong extension mix added. Repeat biochemistry. MassARRAY Analyzer may need calibrating. Contact SEQUENOM Customer Support. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 54 Appendix C Troubleshooting Table C.1 Troubleshooting (continued) Problem Possible Reason Possible Solution Incorrect genotype call leads to incorrect scoring for matches or mismatches. Sample quantity may be too low. Check that sample passed DNA quality control, i.e., had at least 500 amplifiable copies. Incorrect reagent additions. Delete plate from Sample ID database, use Typer to correct the genotype call, and re-run the Sample ID analysis on that plate; and/or repeat experiment if reagent additions were incorrect. One sample has a poor spectra. Manually inspect the affected spectra and re-dispense the affected sample. Unexpected mismatch between a paired normal and tumor sample. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 Appendix D Licensing License. Sequenom's patented nucleic acid analysis by mass spectrometry methods (including iPLEX ®, SensiPLEX TM, OncoCartaTM, and MassCLEAVE TM methods and assays) are protected under United States patent rights including but not limited to, 6,500,621, 6,300,076, 6,258,538, 5,869,242, 6,238,871, 6,440,705, 6,994,969, 7,019,288, 7,419,787, 7,390,672, 7,888,127, and 8,003,317 patents pending including but not limited to 20040081993A1, 11/089,805, and all of the foreign equivalent patent rights of the foregoing. With the purchase of specially designated Sequenom SpectroCHIP ® chips and reagent kit products as set forth in Table D.1. Customer is granted a limited right to practice Sequenom's patented iPLEX ®, OncoCartaTM, ADME PGx, and MassCLEAVE TM methods for certain Licensed Fields of Use as set forth in Table D.1, and subject to the commercial terms which will be provided separately. The licensed rights are not transferable, are for the benefit of Customer only, and expire with the consumption of the SpectroCHIP ® chips purchased. Transfer or resale of products and their components, purchased from Sequenom is prohibited. The license rights granted are limited to one-time use only for each element per SpectroCHIP ® chip purchased. For example, Sequenom's SpectroCHIP ® 384 chip is provided with 384 elements and each element may be used only once. Remanufacture or re-use of Sequenom's SpectroCHIP ® chips and/or elements in conjunction with performing Sequenom's patented nucleic acid analysis methods, is prohibited. Reverse engineering Sequenom products is prohibited. Except as expressly licensed with the purchase of chips and reagent kit products as set forth herein, Sequenom reserves all rights and no additional license rights are granted or implied. For the avoidance of doubt, license rights to patent rights owned or controlled by Sequenom related to analysis of specific biomarkers or genetic regions and/or related to analysis of fetal nucleic acids, are not granted pursuant to these terms and conditions, are not granted with the purchase or license of Sequenom products, and must be separately and independently licensed from Sequenom under a separate written agreement. Table D.1 Sequenom Kits, Applications, and Licensing Sequenom Kit 1. ® iPLEX reagents and SpectroCHIP ® chips 2. OncoCartaTM reagents and SpectroCHIP® chips 3. ADME PGx reagents and SpectroCHIP chips MassCLEAVE TM reagents and SpectroCHIP ® chips Application Licensed Field of Use With Kit Purchase • Genotyping Internal research and development, clinical trial research, for-profit clinical trial support, and for-profit research services that do not require CLIA* registration or licensure (or regional equivalent for Customers outside the United States), by mass spectrometry, and specifically excluding Diagnostic use and Laboratory Developed (a.k.a home-brew) Test use** • QGE quantitative gene expression analysis • SNP Discovery Internal research and development, clinical TM • EpiTYPER methylation research, for-profit clinical study support, and pattern analysis • iSEQ TM comparative sequence analysis and microbial and viral identification and typing for-profit research services that do not require CLIA* registration or licensure (or regional equivalent for Customers outside the United States), by mass spectrometry, and specifically excluding diagnostic and prognostic use and use in the development of Laboratory Developed Tests** *(Clinical Laboratory Improvement Amendments, 1988) ** "Diagnostic use" and Prognostic Use" and "Laboratory Developed Test use" include within their meanings, but are not limited to, providing a service, information, or data in conjunction with diagnostic, prognostic, predictive, therapeutic, prophylactic, or pharmacogenetic testing, determination, or analysis on human subjects, and further include diagnosing or monitoring any disease or its sequelae, state of health, or condition in humans, and testing for treatment selection or response to treatment for any disease, state, or conditions in humans. Software Licenses. For purposes of this Agreement, "Software" means computer software or programs supplied by Sequenom to Customer including but not limited to such software iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 56 Appendix D Licensing that is embedded in or forms an integral part of Sequenom's hardware products in addition to separately provided software for application specific purposes. For purposes of this Agreement, Software does not include Oracle Corporation software products provided by Sequenom as such Oracle products are separately governed by the attached Oracle End User License Terms. (a) Sequenom grants Customer a non-exclusive and non-transferable license to use the Software for processing data for the applicable Licensed Field of Use(s) as set forth in Table D.1 above, and subject to the commercial terms which will be provided separately. Customer shall not permit another party to use the Software and Customer shall effect and maintain adequate security measures to safeguard the Software from access or use by unauthorized persons. Customer shall not transfer, rent, lease, sublicense, loan, copy, modify, adapt, merge, translate, reverse engineer, decompile, or disassemble the Software or create derivative works based on the whole or any part of the Software. (b) The Software license shall not be deemed to extend to any of Sequenom's intellectual property rights, including rights in source code. No copies may be made of the Software without the prior written consent of Sequenom, except that Customer may make a single back up or archival copy. The Software license shall apply to any copy as it applies to the Software. (c) With specific reference to Sequenom's iPLEX ® application Software (the "iPLEX ® Software") that is part of Sequenom's Typer Software product, Customer is provided a non-exclusive and non-transferable license to use the iPLEX® Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such iPLEX ® Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and iPLEX® reagent kit products purchased from Sequenom. License rights to use the iPLEX® Software are not granted by or with the iPLEX ® Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. (d) With specific reference to Sequenom's MassARRAY® QGE Analyzer Software, Customer is provided a non-exclusive and non-transferable license to use the QGE Analyzer Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such QGE Analyzer Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and iPLEX ® reagent kit products purchased from Sequenom. License rights to use the QGE Analyzer Software are not granted by or with the QGE Analyzer Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. (e) With specific reference to Sequenom's SNP Discovery Software, Customer is provided a non-exclusive and non-transferable license to use the SNP Discovery Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such SNP Discovery Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and MassCLEAVETM reagent kit products purchased from Sequenom. License rights to use the SNP Discovery Software are not granted by or with the SNP Discovery Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. (f) With specific reference to Sequenom's EpiTYPERTM Software, Customer is provided a non-exclusive and non-transferable license to use the EpiTYPER TM Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such EpiTYPERTM Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and MassCLEAVE TM reagent kit products purchased from Sequenom. License rights to use the EpiTYPER TM Software are not granted by or with the EpiTYPER TM Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 57 (g) With specific reference to Sequenom's iSEQTM Software, Customer is provided a non-exclusive and non-transferable license to use the iSEQ TM Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such iSEQ TM Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and MassCLEAVE TM reagent kit products purchased from Sequenom. License rights to use the iSEQ TM Software are not granted by or with the iSEQ TM Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. (h) With specific reference to Sequenom's OncoCarta TM Software, Customer is provided a non-exclusive and non-transferable license to use the OncoCarta TM Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such OncoCarta TM Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and OncoCarta TM reagent kit products purchased from Sequenom. License rights to use the OncoCartaTM Software are not granted by or with the OncoCarta TM Software by itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom. (i) With specific reference to Sequenom’s ADME PGx Software, Customer is provided a royalty-free, non-exclusive and non-transferable license to use the ADME PGx Software subject to the terms and conditions in subsections (a) and (b) above, solely upon the condition that such ADME PGx Software is used exclusively in conjunction with specially designated SpectroCHIP ® chips and ADME PGx reagent kit itself, separate and apart from use in conjunction with the specially designated kit products purchased from Sequenom All Software licenses shall terminate automatically and immediately if Customer fails to abide by any of the terms and conditions of this Agreement. Except as expressly licensed herein, Sequenom reserves all rights. Other than as expressly set forth herein, no license rights are granted or implied. No Governmental Approval. Customer is hereby put on notice that all of Sequenom's products and Software currently available as Research Use Only have not been subjected to regulatory review or cleared or approved by the United States Food and Drug Administration, by any other United States governmental agency or entity, or by any equivalent or similar governmental agency or entity outside the United States, and are not CLIA (or the regional equivalent thereof) registered or licensed or otherwise registered, licensed, or approved under any statute, rule, law, or regulation, for any purpose, research, commercial, diagnostic, medical, or otherwise. Customer bears sole responsibility and liability for validating all products purchased by Customer for Customer's intended use. ORACLE END USER LICENSE TERMS ORACLE CORPORATION ("ORACLE") SOFTWARE PRODUCTS (a) The Customer shall limit its use of Oracle's products to the scope of the application package and to its business operations; (b) The Customer shall not transfer Oracle's products except for temporary transfer in the event of computer malfunction; (c) The Customer shall not assign Oracle's products or any interest in Oracle's products. If a security interest is granted in Oracle's products, the secured party has no right to use or transfer Oracle's products; iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050 58 Appendix D Licensing (d) The Customer shall not operate a timeshare, service bureau, subscription or rental use of Oracle's products; (e) Title to Oracle's products remains with Oracle and shall not pass to the Customer or to any other party; (f) The Customer shall not reverse engineer, disassemble or decompile Oracle's products unless required for interoperability and then only to the extent so required for such interoperability; (g) The Customer shall not duplicate Oracle's products except for a sufficient number of copies for the Customer's licensed use and a single back up or archival copy; (h) Oracle shall not be liable for any damages whether indirect, incidental or consequential arising from the use of its products; (i) Where the licence which is granted by Sequenom in respect of Oracle products expires or terminates and is not renewed, the Customer shall discontinue use and destroy or return all copies of Oracle's products and documentation to Sequenom; (j) The Customer shall not cause to be publicised any results of benchmark tests run on Oracle's products; (k) The Customer shall comply fully with all relevant export laws and regulations of the United States and other applicable export and import laws to assure that neither the Oracle products themselves nor any direct products thereof are exported, directly or indirectly in violation of applicable laws; (l) Oracle is not required to perform any obligations other than to the extent agreed by Sequenom and Oracle; (m) Sequenom has the right to audit Customer's use of Oracle's products and report such use to Oracle, or to assign such right to another person; (n) Oracle is a third party beneficiary of these Customer license terms; (o) The Uniform Computer Information Transactions Act shall not apply; and (p) To the extent that Oracle source code is included with any application package, such source code is similarly governed by the terms above. iPLEX® Pro Sample ID Panel User Guide SQNM-USG-CUS-050
© Copyright 2026