Run Quick Sort on Components for the string S = âabracadabra
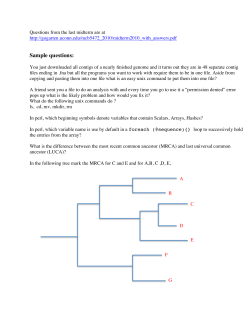
CSCI-598 Spring 2015 Homework 2 Part 1. Paper and pencil. Submit in class. Run Quick Sort on Components for the string S = “abracadabra$”. Outline of this algorithm as well as an example are provided on Blackboard. Part 2. Programming Assignment. Submit on Blackboard. Given three files genome.txt, suffix_array.txt and reads.txt, map reads to the reference genome using the given suffix array, i.e. for each read find a unique position in the genome, where this read exactly matches. Do not output the results for ambiguous reads (a read that is mapped to two or more locations including different strands, +/-, is called ambiguous). Input format of genome.txt: Each line is a 60 characters long string (except the last line in the file might be a shorter string). Input format of suffix_array.txt: the file contains integers separated by a white space, and at the end of all integers there is a new line. The integers are given in the order they occur in the suffix array built for the given genome, i.e. store the first integer at suffix_array[0] (where suffix_array is an array of length = genome size + 1), the second integer at suffix_array[1] and so on. Input format of reads.txt: Each line represents a single read of length 32 characters. 1. Read in genome and append $ sign at the end of the genome. Genome’s size is 6998 and after appending genome’s size is 6999. 2. Read in integers from suffix_array.txt and store them in the array of size equal to the genome’s size (6999). 3. For each read, calculate Lw and Rw (as was explained in lecture slides; you have to modify code to calculate Rw), the lowest and highest indices of suffix_array, that contain positions of suffixes of the genome that start with the given read. 4. Take a reverse-complement of the read and calculate Lw and Rw for the reverse complement. 5. Output the results in the same format as in HW1, where the position in the genome, to which the read maps is given by suffix_array[Lw] Example: for genome AATATACGATATATCGACGGATATG$, the suffix array is: 25 0 5 16 3 1 8 10 20 12 22 14 6 17 24 15 7 Output format: Read_ID read position_in_genome strand(+ or -) Where Read_ID is the order of the read in the input file (starting with 1). Submission on the Blackboard: 1. Submit your zipped code. 2. Submit your output file as a text file or PDF file. 19 18 4 2 9 11 21 13 23
© Copyright 2026